We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1067
From Proteopedia
(Difference between revisions)
| Line 2: | Line 2: | ||
== Zinc Transporter Yiip == | == Zinc Transporter Yiip == | ||
| - | + | ||
== Structure == | == Structure == | ||
| - | + | Yiip's monomer structure is classified into two different domains, the Trans-Membrane Domain (TMD) and C-terminus Domain (CTD). The TMD consists of six helices, forming binding site A, four of which are oriented in a parallel manner with respect to each other, while the remaining two are anti parallel to this four helix cluster. A "salt bridge" or "charge interlock" consisting of four amino acid residues, two Lysines and two Aspartates, forms a junction that both monomers of Yiip converge at, forming a pivot point for protein conformation changes. | |
| - | + | ||
== Salt Bridge == | == Salt Bridge == | ||
== Electrostatic Interactions == | == Electrostatic Interactions == | ||
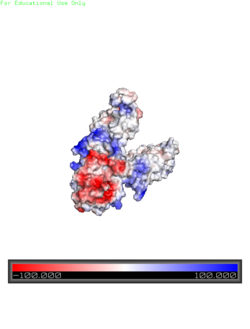
| - | <scene name='69/694234/Electrostatic/1'>Charge distribution</scene> along the exterior surface of the protein is primarily neutral for the | + | <scene name='69/694234/Electrostatic/1'>Charge distribution</scene> along the exterior surface of the protein is primarily neutral for the TMDs, but transitions to positive near the location of the charge interlock and interior side of the cell membrane. This positive section is characteristic of trans-membrane proteins as a means of achieving proper orientation. Binding sites A, B, and C, as well as the CTDs of both monomers, all possess a high negative charge relative to the other charges present, facilitating the binding and releasing of Zn<sup>2+</sup> ions. The two CTDs are held together by the charge interlock and hydrophobic interactions of the TMDs despite their electrostatic repulsion. Upon the release of Zn<sup>2+</sup> ions, the CTDs undergo electronegativity alterations, forcing the two domains apart. [[Image:Yiip_Electrostatic.png|250px|left|thumb|Electrostatics]] |
== Conformation Changes == | == Conformation Changes == | ||
| - | |||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:50, 17 March 2017
|
Contents |
Zinc Transporter Yiip
Structure
Yiip's monomer structure is classified into two different domains, the Trans-Membrane Domain (TMD) and C-terminus Domain (CTD). The TMD consists of six helices, forming binding site A, four of which are oriented in a parallel manner with respect to each other, while the remaining two are anti parallel to this four helix cluster. A "salt bridge" or "charge interlock" consisting of four amino acid residues, two Lysines and two Aspartates, forms a junction that both monomers of Yiip converge at, forming a pivot point for protein conformation changes.
Salt Bridge
Electrostatic Interactions
along the exterior surface of the protein is primarily neutral for the TMDs, but transitions to positive near the location of the charge interlock and interior side of the cell membrane. This positive section is characteristic of trans-membrane proteins as a means of achieving proper orientation. Binding sites A, B, and C, as well as the CTDs of both monomers, all possess a high negative charge relative to the other charges present, facilitating the binding and releasing of Zn2+ ions. The two CTDs are held together by the charge interlock and hydrophobic interactions of the TMDs despite their electrostatic repulsion. Upon the release of Zn2+ ions, the CTDs undergo electronegativity alterations, forcing the two domains apart.Conformation Changes
</StructureSection>