We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
Officially defined as residues 19-90. | Officially defined as residues 19-90. | ||
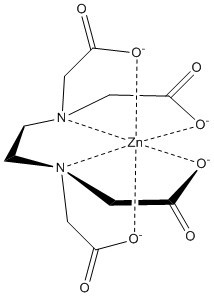
====3His/1Cys Motif==== | ====3His/1Cys Motif==== | ||
| + | |||
| + | [[Image:Zinc_coordinated_chemdraw.jpg]] | ||
== Mechanism of Action == | == Mechanism of Action == | ||
<scene name='69/694239/Hydrophobicity_int_residues/3'>Hydrophobic residues</scene> on the alpha helices of the zinc binding domain participate in van der waals interactions that induce a conformational change in the protein when zinc binds. When Zinc binds, the <scene name='69/694239/Czbd_with_helices_labeled/2'>𝝰1 helix</scene> becomes straightened, burying nonpolar side-chain residues, influencing activity of DgcZ | <scene name='69/694239/Hydrophobicity_int_residues/3'>Hydrophobic residues</scene> on the alpha helices of the zinc binding domain participate in van der waals interactions that induce a conformational change in the protein when zinc binds. When Zinc binds, the <scene name='69/694239/Czbd_with_helices_labeled/2'>𝝰1 helix</scene> becomes straightened, burying nonpolar side-chain residues, influencing activity of DgcZ | ||
Revision as of 03:19, 31 March 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Diguanylate Cyclase DgcZ from Escherichia coli
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644