We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 29: | Line 29: | ||
=== Zinc Binding Site === | === Zinc Binding Site === | ||
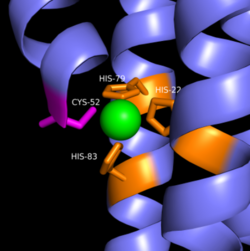
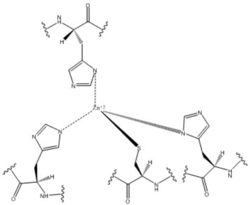
Most cells possess efficient Zinc uptake systems, as Zinc is a reactive Lewis Acid. Zinc binds incredibly tightly to this enzyme at subfemtomolar concentrations. The Zinc co-purified with the protein.Zinc allosterically inhibits the activity of enzyme DgcZ through two allosteric binding sites located on the CZB domain. The inhibition prevents regulation of GGDEF domain function, the location of the active site. The CZB domain is folded into four anti-parallel α-helices as a 2-fold symmetric homodimer, with the N-terminus on the helix 𝝰4. The allosteric binding site includes a <scene name='69/694239/Zinc_binding_domain/4'>3His/1Cys</scene> motif that uses amino acids, H22 of 𝝰1, C52 of 𝝰2, and H79 and H83 of 𝝰3, that span three of the four alpha helices of the CZB domain coordinating the Zinc residue in a tetrahedral fashion. Zahringer et al. mutated Cys52 to Ala through <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html site-directed mutagenesis]</span>, resulting in a lack of coordination on α2. The cysteine residue is not essential for Zinc binding, as Zinc still coordinates to the three His residues with the Cys52Ala mutation, but α2 is free to move and expose the Zinc binding pocket. This exposure was found to lower the protein's affinity for zinc, as the mutation of cysteine to alanine increased the activity of the DgcZ. Using EDTA, Zinc can be removed from the CZB domain. The zinc has higher affinity for EDTA than CZB when EDTA concentration is higher than the concentration of DgcZ. When not coordinated to zinc, the CZB domain adopts a conformation that straightens the <scene name='69/694239/Czbd_with_helices_labeled/2'>𝝰1 helix</scene>, shifting <scene name='69/694239/Hydrophobicity_int_residues/3'>hydrophobic residues</scene> on the α-helices into the center and the GGEEF domain into its productive conformation, increasing activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production. | Most cells possess efficient Zinc uptake systems, as Zinc is a reactive Lewis Acid. Zinc binds incredibly tightly to this enzyme at subfemtomolar concentrations. The Zinc co-purified with the protein.Zinc allosterically inhibits the activity of enzyme DgcZ through two allosteric binding sites located on the CZB domain. The inhibition prevents regulation of GGDEF domain function, the location of the active site. The CZB domain is folded into four anti-parallel α-helices as a 2-fold symmetric homodimer, with the N-terminus on the helix 𝝰4. The allosteric binding site includes a <scene name='69/694239/Zinc_binding_domain/4'>3His/1Cys</scene> motif that uses amino acids, H22 of 𝝰1, C52 of 𝝰2, and H79 and H83 of 𝝰3, that span three of the four alpha helices of the CZB domain coordinating the Zinc residue in a tetrahedral fashion. Zahringer et al. mutated Cys52 to Ala through <span class="plainlinks">[http://www.chem.qmul.ac.uk/iubmb/enzyme/EC3/5/3/18.html site-directed mutagenesis]</span>, resulting in a lack of coordination on α2. The cysteine residue is not essential for Zinc binding, as Zinc still coordinates to the three His residues with the Cys52Ala mutation, but α2 is free to move and expose the Zinc binding pocket. This exposure was found to lower the protein's affinity for zinc, as the mutation of cysteine to alanine increased the activity of the DgcZ. Using EDTA, Zinc can be removed from the CZB domain. The zinc has higher affinity for EDTA than CZB when EDTA concentration is higher than the concentration of DgcZ. When not coordinated to zinc, the CZB domain adopts a conformation that straightens the <scene name='69/694239/Czbd_with_helices_labeled/2'>𝝰1 helix</scene>, shifting <scene name='69/694239/Hydrophobicity_int_residues/3'>hydrophobic residues</scene> on the α-helices into the center and the GGEEF domain into its productive conformation, increasing activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production. | ||
| + | |||
| + | [[Image:Electrostatic map front view CZB.png|250 px|left|thumb|]] | ||
=== Other Ligands === | === Other Ligands === | ||
Revision as of 17:51, 31 March 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Diguanylate Cyclase DgcZ from Escherichia coli
| |||||||||||