Carboxypeptidase A
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
{{Sandbox_Reserved_Butler_CH462_Sp2015_#}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | {{Sandbox_Reserved_Butler_CH462_Sp2015_#}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
=Carboxypeptidase A in ''Bos taurus''= | =Carboxypeptidase A in ''Bos taurus''= | ||
| - | <StructureSection load='1cpx' size='340' side='right' scene='69/694222/1cpx_default/ | + | <StructureSection load='1cpx' size='340' side='right' scene='69/694222/1cpx_default/2'> |
==Introduction== | ==Introduction== | ||
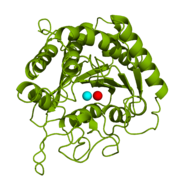
[[Image:1cpx - 2 zinc ions.png|thumb|Figure 1: Catalytic and inhibitory Zn<sup>2+</sup> ions in the active site of 1CPX. The catalytic and inhibitory Zn<sup>2+</sup> ions are shown in cyan and red, respectively. PDB code: [http://www.rcsb.org/pdb/explore/explore.do?structureId=1cpx 1CPX].]] | [[Image:1cpx - 2 zinc ions.png|thumb|Figure 1: Catalytic and inhibitory Zn<sup>2+</sup> ions in the active site of 1CPX. The catalytic and inhibitory Zn<sup>2+</sup> ions are shown in cyan and red, respectively. PDB code: [http://www.rcsb.org/pdb/explore/explore.do?structureId=1cpx 1CPX].]] | ||
| - | <scene name='69/694222/1cpx_default/ | + | <scene name='69/694222/1cpx_default/2'>Carboxypeptidase A (peptidyl-L-amino acid hydrolase, EC 3.4.17.1, often abbreviated CPA)</scene> is a metallo[http://en.wikipedia.org/wiki/Exopeptidase exopeptidase] whose biological function is to cleave the [http://en.wikipedia.org/wiki/C-terminus C-terminal] amino acid residue from polypeptide substrates.<ref name="CPA1">Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. ''Biochemistry''. 37(47):16555-16564. [http://pubs.acs.org/doi/abs/10.1021/bi981678i DOI: 10.1021/bi981678i]</ref> Specifically, CPA is one member of a large group of Zn<sup>2+</sup> [http://en.wikipedia.org/wiki/Metalloprotein#Metalloenzymes metalloenzymes] that carries out the hydrolysis of C-terminal polypeptide residues through the [http://en.wikipedia.org/wiki/Deprotonation deprotonation] of a water molecule that is coordinated to the Zn<sup>2+</sup> ion in the enzyme's [http://en.wikipedia.org/wiki/Active_site active site].<ref name="CPA2">Christianson DW, Lipscomb WN. 1989. Carboxypeptidase A. ''Acc. Chem. Res.'' 22:62-69.</ref> CPA consists of a single polypeptide chain that contains 307 amino acids. Produced in the pancreas, CPA itself must first be modified by [http://en.wikipedia.org/wiki/Trypsin trypsin] and [http://en.wikipedia.org/wiki/Chymotrypsin chymotrypsin] in order to achieve an active form that serves its biological function.<ref name="CPA1" /> Although different biologically active forms of CPA are found across different species, including humans, much research has investigated bovine pancreatic zinc carboxypeptidase A. [http://en.wikipedia.org/wiki/X-ray_crystallography X-ray crystallography] has demonstrated that bovine CPA has the ability to bind two Zn<sup>2+</sup> ions in its active site, in which the binding of one Zn<sup>2+</sup> is catalytic (shown in cyan), while the binding of a second Zn<sup>2+</sup> inhibits the hydrolysis reaction mechanism (shown in red) (Figure 1).<ref name="CPA1" /> An example of a crystal structure for active CPA (one zinc bound) has been deposited in the [http://www.rcsb.org/pdb/home/home.do Protein Data Bank (PDB) database] as [http://www.rcsb.org/pdb/explore/explore.do?structureId=3cpa 3CPA]. An inhibited CPA (two zincs bound) has been deposited under the label [http://www.rcsb.org/pdb/explore/explore.do?structureId=1cpx 1CPX]. |
==Structure== | ==Structure== | ||
| - | Bovine CPA exists as a single unit with [http://en.wikipedia.org/wiki/Molecular_symmetry C1 symmetry] in the pancreatic physiological environment. According to [http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3CPA structural information] deposited in the PDB database, the single polypeptide chain of CPA contains a mixture of <scene name='69/694222/3cpasecondarystructure/1'>α-helices and β-sheets</scene>, of which there are a total of 11 helices (one [http://en.wikipedia.org/wiki/310_helix 3<sub>10</sub>], eight [http://en.wikipedia.org/wiki/Alpha_helix 3.6<sub>13</sub>]) and ten [http://en.wikipedia.org/wiki/Beta_sheet β-strands]. The helices are shown in magenta, and the β-strands are displayed in yellow. A [http://en.wikibooks.org/wiki/Structural_Biochemistry/Chemical_Bonding/_Disulfide_bonds disulfide bond] connects the residues Cys138 and Cys161. The disulfide bond can be seen in yellow in the <scene name='69/694222/1cpx_default/ | + | Bovine CPA exists as a single unit with [http://en.wikipedia.org/wiki/Molecular_symmetry C1 symmetry] in the pancreatic physiological environment. According to [http://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=3CPA structural information] deposited in the PDB database, the single polypeptide chain of CPA contains a mixture of <scene name='69/694222/3cpasecondarystructure/1'>α-helices and β-sheets</scene>, of which there are a total of 11 helices (one [http://en.wikipedia.org/wiki/310_helix 3<sub>10</sub>], eight [http://en.wikipedia.org/wiki/Alpha_helix 3.6<sub>13</sub>]) and ten [http://en.wikipedia.org/wiki/Beta_sheet β-strands]. The helices are shown in magenta, and the β-strands are displayed in yellow. A [http://en.wikibooks.org/wiki/Structural_Biochemistry/Chemical_Bonding/_Disulfide_bonds disulfide bond] connects the residues Cys138 and Cys161. The disulfide bond can be seen in yellow in the <scene name='69/694222/1cpx_default/2'>original rotating figure</scene>. |
Six different biologically active forms of the CPA monomeric unit exist. <scene name='69/694222/1cpxcleavageforms/2'>Three of these active forms</scene> are produced following the cleavage of amino acid residue segments from the initial [http://en.wikipedia.org/wiki/Zymogen zymogen], or proenzyme, by trypsin and chymotrypsin, which are also found in the pancreas. Cleavage by trypsin generates either the '''α-form''' (residues Ala1-Asn307) or the '''β-form''' (residues Ser3-Asn307). Chymotrypsin cleavage generates the '''γ-form''' (residues Asn8-Asn307). The α-form essentially is the protein without any additional residue cleavages. The Ala and Arg residues, shown in red and white respectively, are cleaved in the β-form. In addition to the red and white residues, the residues displayed in yellow are cleaved to give the γ-form. The <scene name='69/694222/3cpageneticforms/3'>other three active forms</scene> of CPA arise from [http://en.wikipedia.org/wiki/Genetic_variation genetic variation] in residues located at three separate positions of the polypeptide chain. The differences include the following: Ile/Val179, Ala/Glu228, and Val/Leu305.<ref name="CPA1" /> Each of the six biologically active monomeric units carry out the same function of hydrolyzing the C-terminal [http://en.wikipedia.org/wiki/Peptide_bond peptide bond] of a polypeptide substrate. | Six different biologically active forms of the CPA monomeric unit exist. <scene name='69/694222/1cpxcleavageforms/2'>Three of these active forms</scene> are produced following the cleavage of amino acid residue segments from the initial [http://en.wikipedia.org/wiki/Zymogen zymogen], or proenzyme, by trypsin and chymotrypsin, which are also found in the pancreas. Cleavage by trypsin generates either the '''α-form''' (residues Ala1-Asn307) or the '''β-form''' (residues Ser3-Asn307). Chymotrypsin cleavage generates the '''γ-form''' (residues Asn8-Asn307). The α-form essentially is the protein without any additional residue cleavages. The Ala and Arg residues, shown in red and white respectively, are cleaved in the β-form. In addition to the red and white residues, the residues displayed in yellow are cleaved to give the γ-form. The <scene name='69/694222/3cpageneticforms/3'>other three active forms</scene> of CPA arise from [http://en.wikipedia.org/wiki/Genetic_variation genetic variation] in residues located at three separate positions of the polypeptide chain. The differences include the following: Ile/Val179, Ala/Glu228, and Val/Leu305.<ref name="CPA1" /> Each of the six biologically active monomeric units carry out the same function of hydrolyzing the C-terminal [http://en.wikipedia.org/wiki/Peptide_bond peptide bond] of a polypeptide substrate. | ||
Revision as of 19:16, 14 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Carboxypeptidase A in Bos taurus
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. Biochemistry. 37(47):16555-16564. DOI: 10.1021/bi981678i
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Christianson DW, Lipscomb WN. 1989. Carboxypeptidase A. Acc. Chem. Res. 22:62-69.
- ↑ Suh J, Cho W, Chung S. 1985. Carboxypeptidase A-catalyzed hydrolysis of α-(acylamino)cinnamoyl derivatives of L-β-phenyllactate and L-phenylalaninate: evidence for acyl-enzyme intermediates. J. Am. Chem. Soc. 107:4530-4535. DOI: 10.1021/ja00301a025
- ↑ Geoghegan, KF, Galdes, A, Martinelli, RA, Holmquist, B, Auld, DS, Vallee, BL. 1983. Cryospectroscopy of intermediates in the mechanism of carboxypeptidase A. Biochem. 22(9):2255-2262. DOI: 10.1021/bi00278a031
- ↑ Kaplan, AP, Bartlett, PA. 1991. Synthesis and evaluation of an inhibitor of carboxypeptidase A with a Ki value in the femtomolar range. Biochem. 30(33):8165-8170. PMID: 1868091
- ↑ Worthington, K., Worthington, V. 1993. Worthington Enzyme Manual: Enzymes and Related Biochemicals. Freehold (NJ): Worthington Biochemical Corporation; [2011; accessed March 28, 2017]. Carboxypeptidase A. http://www.worthington-biochem.com/COA/
- ↑ Pitout, MJ, Nel, W. 1969. The inhibitory effect of ochratoxin a on bovine carboxypeptidase a in vitro. Biochem. Pharma. 18(8):1837-1843. DOI: 0.1016/0006-2952(69)90279-2
- ↑ Normant, E, Martres, MP, Schwartz, JC, Gros, C. 1995. Purification, cDNA cloning, functional expression, and characterization of a 26-kDa endogenous mammalian carboxypeptidase inhibitor. Proc. Natl. Acad. Sci. 92(26):12225-12229. PMCID: PMC40329
Student Contributors
- Thomas Baldwin
- Michael Melbardis
- Clay Schnell
Proteopedia Page Contributors and Editors (what is this?)
Michael Melbardis, Douglas Schnell, Thomas Baldwin, Geoffrey C. Hoops, Michal Harel