Carboxypeptidase A
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
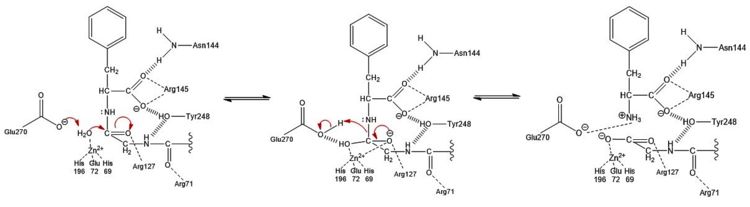
The second mechanism, which has been coined as the promoted water pathway, is better supported by chemical and structural data. The mechanism of the reaction (Figure 3) is as follows: | The second mechanism, which has been coined as the promoted water pathway, is better supported by chemical and structural data. The mechanism of the reaction (Figure 3) is as follows: | ||
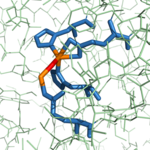
| - | # Three S1 subsite residues (Asn144, Arg145, and Tyr248) and the hydrophobic S1' subsite recognize the C-terminus of the polypeptide substrate. | + | # Three S1 subsite residues (<scene name='69/694222/3cpas1subsiteresidues1/2'>Asn144, Arg145, and Tyr248</scene>) and the <scene name='69/694222/3cpahydrophobicpocketresidues/3'>hydrophobic S1' subsite</scene> recognize the C-terminus of the polypeptide substrate. |
| - | # After aiding in the recognition of the substrate, Tyr248 "caps" the binding pocket. | + | # After aiding in the recognition of the substrate, <scene name='69/694222/3cpadeeppocket2/1'>Tyr248 "caps" the binding pocket</scene>. |
# The catalytic Zn<sup>2+</sup> ion and Arg127 residue engage in ion-dipole interactions with the oxygen atom of the carbonyl group of the C-terminal peptide bond, further polarizing the carbon to oxygen double bond. | # The catalytic Zn<sup>2+</sup> ion and Arg127 residue engage in ion-dipole interactions with the oxygen atom of the carbonyl group of the C-terminal peptide bond, further polarizing the carbon to oxygen double bond. | ||
# A water molecule, which has been [http://en.wikipedia.org/wiki/Deprotonation deprotonated] by Glu270 ([http://bio.libretexts.org/Core/Biochemistry/Catalysis/METHODS_OF_CATALYSIS/General_Acid%2F%2FBase_Catalysis base catalyst]) and is being held in place one bond distance away from the partially positive carbon of the C-terminal carbonyl, acts as a [http://en.wikipedia.org/wiki/Nucleophile nucleophile] and attacks this carbon to generate a [http://goldbook.iupac.org/T06289.html tetrahedral intermediate] stabilized by both the Zn<sup>2+</sup> ion and surrounding positive charges of S1 subsite residues. | # A water molecule, which has been [http://en.wikipedia.org/wiki/Deprotonation deprotonated] by Glu270 ([http://bio.libretexts.org/Core/Biochemistry/Catalysis/METHODS_OF_CATALYSIS/General_Acid%2F%2FBase_Catalysis base catalyst]) and is being held in place one bond distance away from the partially positive carbon of the C-terminal carbonyl, acts as a [http://en.wikipedia.org/wiki/Nucleophile nucleophile] and attacks this carbon to generate a [http://goldbook.iupac.org/T06289.html tetrahedral intermediate] stabilized by both the Zn<sup>2+</sup> ion and surrounding positive charges of S1 subsite residues. | ||
# The peptide bond is cleaved through an addition-elimination step. | # The peptide bond is cleaved through an addition-elimination step. | ||
| - | # The Glu270 base catalyst is regenerated through a final [http://www.masterorganicchemistry.com/tips/proton-transfer/ proton transfer] with the nitrogen atom of the former C-terminal peptide bond. | + | # The <scene name='69/694222/3cpas1subsiteglu270/2'>Glu270 base catalyst</scene> is regenerated through a final [http://www.masterorganicchemistry.com/tips/proton-transfer/ proton transfer] with the nitrogen atom of the former C-terminal peptide bond. |
# Product release is facilitated, in part, by unfavorable electrostatic interactions between the regenerated Glu270 base catalyst and the deprotonated carboxylic acid at the new C-terminus. | # Product release is facilitated, in part, by unfavorable electrostatic interactions between the regenerated Glu270 base catalyst and the deprotonated carboxylic acid at the new C-terminus. | ||
[[Image:Proteopedia Reaction Mechanism Graphic.jpg|750 px|thumb|left|Figure 3: Hydrolysis of C-terminal polypeptide substrate residue by CPA using the promoted water pathway. Residues of the S1 subsite stabilize the negatively charged intermediate once the water molecule complexed with the Zn<sup>2+</sup> ion is deprotonated by the base catalyst, Glu270, and attacks the carbonyl. This figure is derived from Figure 10 in "Carboxypeptidase A" by Christianson and Lipscomb (''Acc. Chem. Res.'', 1989).<ref name="CPA2" /> ]] | [[Image:Proteopedia Reaction Mechanism Graphic.jpg|750 px|thumb|left|Figure 3: Hydrolysis of C-terminal polypeptide substrate residue by CPA using the promoted water pathway. Residues of the S1 subsite stabilize the negatively charged intermediate once the water molecule complexed with the Zn<sup>2+</sup> ion is deprotonated by the base catalyst, Glu270, and attacks the carbonyl. This figure is derived from Figure 10 in "Carboxypeptidase A" by Christianson and Lipscomb (''Acc. Chem. Res.'', 1989).<ref name="CPA2" /> ]] | ||
Revision as of 12:21, 18 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
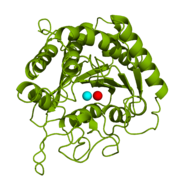
Carboxypeptidase A in Bos taurus
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 Bukrinsky JT, Bjerrum MJ, Kadziola A. 1998. Native carboxypeptidase A in a new crystal environment reveals a different conformation of the important tyrosine 248. Biochemistry. 37(47):16555-16564. DOI: 10.1021/bi981678i
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Christianson DW, Lipscomb WN. 1989. Carboxypeptidase A. Acc. Chem. Res. 22:62-69.
- ↑ Suh J, Cho W, Chung S. 1985. Carboxypeptidase A-catalyzed hydrolysis of α-(acylamino)cinnamoyl derivatives of L-β-phenyllactate and L-phenylalaninate: evidence for acyl-enzyme intermediates. J. Am. Chem. Soc. 107:4530-4535. DOI: 10.1021/ja00301a025
- ↑ Geoghegan, KF, Galdes, A, Martinelli, RA, Holmquist, B, Auld, DS, Vallee, BL. 1983. Cryospectroscopy of intermediates in the mechanism of carboxypeptidase A. Biochem. 22(9):2255-2262. DOI: 10.1021/bi00278a031
- ↑ Kaplan, AP, Bartlett, PA. 1991. Synthesis and evaluation of an inhibitor of carboxypeptidase A with a Ki value in the femtomolar range. Biochem. 30(33):8165-8170. PMID: 1868091
- ↑ Worthington, K., Worthington, V. 1993. Worthington Enzyme Manual: Enzymes and Related Biochemicals. Freehold (NJ): Worthington Biochemical Corporation; [2011; accessed March 28, 2017]. Carboxypeptidase A. http://www.worthington-biochem.com/COA/
- ↑ Pitout, MJ, Nel, W. 1969. The inhibitory effect of ochratoxin a on bovine carboxypeptidase a in vitro. Biochem. Pharma. 18(8):1837-1843. DOI: 0.1016/0006-2952(69)90279-2
- ↑ Normant, E, Martres, MP, Schwartz, JC, Gros, C. 1995. Purification, cDNA cloning, functional expression, and characterization of a 26-kDa endogenous mammalian carboxypeptidase inhibitor. Proc. Natl. Acad. Sci. 92(26):12225-12229. PMCID: PMC40329
Student Contributors
- Thomas Baldwin
- Michael Melbardis
- Clay Schnell
Proteopedia Page Contributors and Editors (what is this?)
Michael Melbardis, Douglas Schnell, Thomas Baldwin, Geoffrey C. Hoops, Michal Harel