User:Natalie Van Ochten/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 17: | Line 17: | ||
====Lid Region Conservation==== | ====Lid Region Conservation==== | ||
| - | The specific residues in the lid region are different in different organisms <ref name="frey" /> (Figure 2). The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that | + | The specific residues in the lid region are different in different organisms <ref name="frey" /> (Figure 2). The only consistent similarity is a <span class="plainlinks">[https://en.wikipedia.org/wiki/Conserved_sequence conserved]</span> leucine residue in this lid that functions to hydrogen bond with the <span class="plainlinks">[https://en.wikipedia.org/wiki/Ligand ligand]</span> bound to the active site in DDAH-1 but not in DDAH-2 <ref name="rasheed" /> (Figure 2). Different <span class="plainlinks">[https://en.wikipedia.org/wiki/Protein_isoform isoforms]</span> from the same species can have differences in lid regions as well <ref name="frey" />. DDAH-2 has a negatively charged lid while DDAH-1 has a positively charged lid <ref name="frey" />. |
| - | [[Image:Lid Region WebLogo.png|500 px|center|thumb|'''Figure 2.''' WebLogo for the lid region in DDAH-1 of | + | [[Image:Lid Region WebLogo.png|500 px|center|thumb|'''Figure 2.''' WebLogo for the lid region in DDAH-1 of eight different organisms.]] |
===Active Site=== | ===Active Site=== | ||
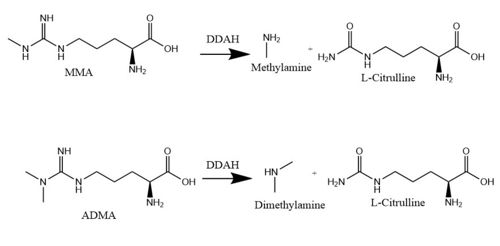
| - | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/4'>Cys249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref> (Figure 3). The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via hydrogen bonds. This is followed by collapsing the tetrahedral product to get rid of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Alkylamines alkylamine]</span> leaving group. A <span class="plainlinks">[https://en.wikipedia.org/wiki/Isothiouronium thiouronium]</span> intermediate is then formed with <span class="plainlinks">[https://en.wikipedia.org/wiki/Orbital_hybridisation sp<sup>2</sup> hybridization]</span>. This intermediate is hydrolyzed to form citrulline. The <scene name='75/752351/Ddah_active_site_his162/2'>His162</scene> protonates the leaving group in this reaction and generates hydroxide to hydrolyze the intermediate formed in the reaction (Figure 3). L-citrulline leaves the active site when the lid opens. The amines can either leave through the entrance to the active site or through a pore made by movement of Glu77 and Lys174 <ref name="frey" />. Studies suggest that Cys249 is neutral until binding of guanidinium near Cys249 decreases Cys249’s <span class="plainlinks">[https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa]</span> and deprotonates the thiolate to activate the nucleophile. Other studies suggest that the Cys249 and an active site His162 form an <span class="plainlinks">[https://en.wikipedia.org/wiki/Intimate_ion_pair ion pair]</span> to deprotonate the thiolate. Cys249 and His162 can also form a binding site for inhibitors to bind to which stabilizes the thiolate. This is important in regulating NO activity in organisms and designing drugs to inhibit this enzyme <ref name="stone" />. | + | The normal DDAH regulation <span class="plainlinks">[https://en.wikipedia.org/wiki/Reaction_mechanism mechanism]</span> depends on the presence of <scene name='75/752351/Ddah_active_site/4'>Cys249</scene> in the active site that acts as a <span class="plainlinks">[https://en.wikipedia.org/wiki/Nucleophile nucleophile]</span> in the mechanism <ref name="stone">Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:<span class="plainlinks">[https://www.ncbi.nlm.nih.gov/pubmed/16634643 16634643]</span> doi:<span class="plainlinks">[http://pubs.acs.org/doi/abs/10.1021/bi052595m 10.1021/bi052595m]</span></ref> (Figure 3). The Cys249 is used to attack the <span class="plainlinks">[https://en.wikipedia.org/wiki/Guanidine guanidinium]</span> carbon on the substrate that is held in the active site via <scene name='75/752351/Hbond_leu29/6'>hydrogen bonds</scene>. This is followed by collapsing the tetrahedral product to get rid of the <span class="plainlinks">[https://en.wikipedia.org/wiki/Alkylamines alkylamine]</span> leaving group. A <span class="plainlinks">[https://en.wikipedia.org/wiki/Isothiouronium thiouronium]</span> intermediate is then formed with <span class="plainlinks">[https://en.wikipedia.org/wiki/Orbital_hybridisation sp<sup>2</sup> hybridization]</span>. This intermediate is hydrolyzed to form citrulline. The <scene name='75/752351/Ddah_active_site_his162/2'>His162</scene> protonates the leaving group in this reaction and generates hydroxide to hydrolyze the intermediate formed in the reaction (Figure 3). L-citrulline leaves the active site when the lid opens. The amines can either leave through the entrance to the active site or through a pore made by movement of Glu77 and Lys174 <ref name="frey" />. Studies suggest that Cys249 is neutral until binding of guanidinium near Cys249 decreases Cys249’s <span class="plainlinks">[https://en.wikipedia.org/wiki/Acid_dissociation_constant pKa]</span> and deprotonates the thiolate to activate the nucleophile. Other studies suggest that the Cys249 and an active site His162 form an <span class="plainlinks">[https://en.wikipedia.org/wiki/Intimate_ion_pair ion pair]</span> to deprotonate the thiolate. Cys249 and His162 can also form a binding site for inhibitors to bind to which stabilizes the thiolate. This is important in regulating NO activity in organisms and designing drugs to inhibit this enzyme <ref name="stone" />. |
[[Image:The Normal DDAH Mechanism.jpg|800px|center|thumb|'''Figure 3.''' The normal mechanism of DDAH highlighting important residues involved.]] | [[Image:The Normal DDAH Mechanism.jpg|800px|center|thumb|'''Figure 3.''' The normal mechanism of DDAH highlighting important residues involved.]] | ||
Revision as of 03:07, 21 April 2017
Dimethylarginine Dimethylaminohydrolase
| |||||||||||
References
- ↑ 1.0 1.1 Palm F, Onozato ML, Luo Z, Wilcox CS. Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. American Journal of Physiology. 2007 Dec 1;293(6):3227-3245. PMID:17933965 doi:10.1152/ajpheart.00998.2007
- ↑ 2.0 2.1 2.2 Tran CTL, Leiper JM, Vallance P. The DDAH/ADMA/NOS pathway. Atherosclerosis Supplements. 2003 Dec;4(4):33-40. PMID:14664901 doi:10.1016/S1567-5688(03)00032-1
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 3.15 3.16 3.17 3.18 3.19 3.20 3.21 3.22 3.23 3.24 Frey D, Braun O, Briand C, Vasak M, Grutter MG. Structure of the mammalian NOS regulator dimethylarginine dimethylaminohydrolase: a basis for the design of specific inhibitors. Structure. 2006 May;14(5):901-911. PMID:[1] doi:10.1016/j.str.2006.03.006
- ↑ Humm A, Fritsche E, Mann K, Göhl M, Huber R. Recombinant expression and isolation of human L-arginine:glycine amidinotransferase and identification of its active-site cysteine residue. Biochemical Journal. 1997 March 15;322(3):771-776. PMID:9148748 doi:10.1042/bj3220771
- ↑ 5.0 5.1 5.2 Rasheed M, Richter C, Chisty LT, Kirkpatrick J, Blackledge M, Webb MR, Driscoll PC. Ligand-dependent dynamics of the active site lid in bacterial Dimethyarginine Dimethylaminohydrolase. Biochemistry. 2014 Feb 18;53:1092-1104. PMCID:PMC3945819 doi:10.1021/bi4015924
- ↑ 6.0 6.1 Stone EM, Costello AL, Tierney DL, Fast W. Substrate-assisted cysteine deprotonation in the mechanism of Dimethylargininase (DDAH) from Pseudomonas aeruginosa. Biochemistry. 2006 May 2;45(17):5618-5630. PMID:16634643 doi:10.1021/bi052595m
- ↑ 7.0 7.1 Pace NJ, Weerpana E. Zinc-binding cysteines: diverse functions and structural motifs. Biomolecules. 2014 June;4(2):419-434. PMCID:4101490 doi:10.3390/biom4020419
- ↑ Janssen W, Pullamsetti SS, Cooke J, Weissmann N, Guenther A, Schermuly RT. The role of dimethylarginine dimethylaminohydrolase (DDAH) in pulmonary fibrosis. The Journal of Pathology. 2012 Dec 12;229(2):242-249. Epub 2013 Jan. PMID:23097221 doi:10.1002/path.4127
- ↑ Colasanti M, Suzuki H. The dual personality of NO. ScienceDirect. 2000 Jul 1;21(7):249-252. PMID:10979862 doi:10.1016/S0165-6147(00)01499-1
- ↑ Rassaf T, Feelisch M, Kelm M. Circulating NO pool: assessment of nitrite and nitroso species in blood and tissues. Free Rad. Biol. Med. 2004 Feb 15;36(4):413-422. PMID:14975444 doi:10.1016/j.freeradbiomed.2003.11.011
- ↑ Tsao PS, Cooke JP. Endothelial alterations in hypercholesterolemia: more than simply vasodilator dysfunction. Journal of Cardiovascular Pharmacology. 1998;32(3):48-53. PMID:9883748
- ↑ Vallance P, Leiper J. Blocking NO synthesis: how, where and why? Nat. Rev. Drug Discov. 2002 Dec;1(12):939-950. PMID:12461516 doi:10.1038/nrd960
Student Contributors
- Natalie Van Ochten
- Kaitlyn Enderle
- Colton Junod
3D Structures of Dimethylarginine Dimethylaminohydrolase
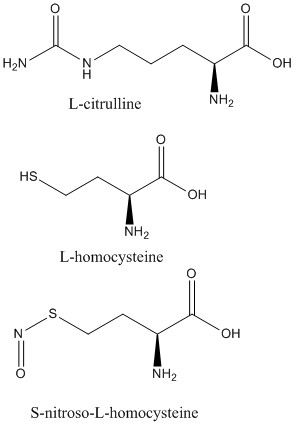
2C6Z L-citrulline bound to isoform 1
2CI1 S-nitroso-L-homocysteine bound to isoform 1
2CI3 crystal form 1
2CI4 crystal form 2
2CI5 L-homocysteine bound to isoform 1
2CI6 Zn (II) bound at low pH to isoform 1
2CI7 Zn (II) bound at high pH to isoform 1