This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
DgcZ is a tetrameric protein from ''E. coli'' made of two domains. Each domain is a dimer, so the whole protein can be called a dimer of dimers. The DgcZ protein has <scene name='69/694239/C2_symmetry/6'>C2</scene> symmetry down its central axis. The catalytic glycine-glycine-glutamate-glutamate-phenylalanine (GGEEF) domain is responsible for synthesizing c-di-GMP, and the regulatory chemoreceptor zinc binding (CZB) domain houses two zinc binding sites. DgcZ binds zinc in the CZB domain with sub-femtomolar (10<sup>-16</sup>M) affinity. When zinc is bound, the CZB and GGEEF domains adopt conformations that inhibit DgcZ function <sup>[1]</sup>. Enzyme DgcZ was co-crystallized with Zinc fixing the structure in its inactivate conformation. The CZB domain is common to many bacterial lineages, including its prevalence in DgcZ homologs. The domain has an important role in signal transduction of bacteria. CZB and GGEEF domains are prevalent in many bacterial proteins from differing strands of ''E. coli'' <sup>[2]</sup>. The GGEEF domain is catalytic in that it contains the active sites used for cyclizing GTP into c-di-GMP. The CZB domain is used for ligand-mediated regulation of c-di-GMP production. | DgcZ is a tetrameric protein from ''E. coli'' made of two domains. Each domain is a dimer, so the whole protein can be called a dimer of dimers. The DgcZ protein has <scene name='69/694239/C2_symmetry/6'>C2</scene> symmetry down its central axis. The catalytic glycine-glycine-glutamate-glutamate-phenylalanine (GGEEF) domain is responsible for synthesizing c-di-GMP, and the regulatory chemoreceptor zinc binding (CZB) domain houses two zinc binding sites. DgcZ binds zinc in the CZB domain with sub-femtomolar (10<sup>-16</sup>M) affinity. When zinc is bound, the CZB and GGEEF domains adopt conformations that inhibit DgcZ function <sup>[1]</sup>. Enzyme DgcZ was co-crystallized with Zinc fixing the structure in its inactivate conformation. The CZB domain is common to many bacterial lineages, including its prevalence in DgcZ homologs. The domain has an important role in signal transduction of bacteria. CZB and GGEEF domains are prevalent in many bacterial proteins from differing strands of ''E. coli'' <sup>[2]</sup>. The GGEEF domain is catalytic in that it contains the active sites used for cyclizing GTP into c-di-GMP. The CZB domain is used for ligand-mediated regulation of c-di-GMP production. | ||
| - | + | ==Catalytic GGEEF Domain=== | |
The <scene name='69/694239/Ggeef_domain_zmout_dgcz/2'>GGEEF domain</scene> of DgcZ is part of the GGDEF family of proteins that includes a conserved sequence, GG[DE][DE]F<sup>[3]</sup>.The GGEEF domain is a homodimer consisting of a central five-stranded β-sheet surrounded by five α-helices. The GGEEF domain contains two catalytic half sites that, when combined together in a productive conformation, form the entire <scene name='69/694239/Ggeef_domain_dgcz/6'>active site</scene>. Residues Gly-206, Gly-207, Glu-208, Glu-209, and Phe-210 form the active site. Each <scene name='69/694239/Ggeef_domain_half_site_dgcz/3'>half-site</scene> binds one GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to <scene name='69/694239/Gtp_guanine_bonds_asn_asp_dgcz/6'>Asn 173 and Asp 182</scene>. This scene depicts an inactive form of the protein because it was crystallized with zinc bound. The ribose of each guanosine triphosphate, and subsequent product c-di-GMP riboses, are held only loosely by the enzyme, while the phosphate groups are not bound to the active site at all<sup>[1]</sup>. | The <scene name='69/694239/Ggeef_domain_zmout_dgcz/2'>GGEEF domain</scene> of DgcZ is part of the GGDEF family of proteins that includes a conserved sequence, GG[DE][DE]F<sup>[3]</sup>.The GGEEF domain is a homodimer consisting of a central five-stranded β-sheet surrounded by five α-helices. The GGEEF domain contains two catalytic half sites that, when combined together in a productive conformation, form the entire <scene name='69/694239/Ggeef_domain_dgcz/6'>active site</scene>. Residues Gly-206, Gly-207, Glu-208, Glu-209, and Phe-210 form the active site. Each <scene name='69/694239/Ggeef_domain_half_site_dgcz/3'>half-site</scene> binds one GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to <scene name='69/694239/Gtp_guanine_bonds_asn_asp_dgcz/6'>Asn 173 and Asp 182</scene>. This scene depicts an inactive form of the protein because it was crystallized with zinc bound. The ribose of each guanosine triphosphate, and subsequent product c-di-GMP riboses, are held only loosely by the enzyme, while the phosphate groups are not bound to the active site at all<sup>[1]</sup>. | ||
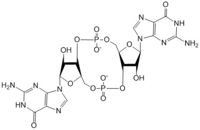
The alpha phosphate is available for attack by the 3 prime hydroxyl group on another GTP. A <scene name='69/694239/Gtp_magnesium_cofactors_dgcz/2'>Magnesium ion</scene> (Mg<sup>2+</sup>) stabilizes the negative charges on the phosphate groups. When in the productive conformation, each GTP is held in close proximity with the α-phosphate groups overlapping C3 of the ribose ring. This conformation allows the α-phospate of one GTP to react with the alcohol group attached to C3 of the ribose on the second GTP, resulting in a cyclization of the two molecules into c-di-GMP. | The alpha phosphate is available for attack by the 3 prime hydroxyl group on another GTP. A <scene name='69/694239/Gtp_magnesium_cofactors_dgcz/2'>Magnesium ion</scene> (Mg<sup>2+</sup>) stabilizes the negative charges on the phosphate groups. When in the productive conformation, each GTP is held in close proximity with the α-phosphate groups overlapping C3 of the ribose ring. This conformation allows the α-phospate of one GTP to react with the alcohol group attached to C3 of the ribose on the second GTP, resulting in a cyclization of the two molecules into c-di-GMP. | ||
| - | + | ===Mechanism of Action=== | |
Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap. | Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap. | ||
| Line 28: | Line 28: | ||
4. The result of this reaction is the C3 alcohol group of each ribose covalently bonded to an α-phosphate forming c-di-GMP. | 4. The result of this reaction is the C3 alcohol group of each ribose covalently bonded to an α-phosphate forming c-di-GMP. | ||
| - | + | ==CZB Domain and Zinc Binding Site== | |
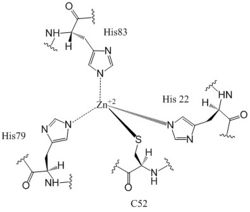
The <scene name='69/694239/Zb_domain_residues_19-90/5'>CZB Domain</scene>, residues 19-90, is responsible for regulating the function of DgcZ due to the presence of two zinc binding sites. The domain contains the <scene name='69/694239/Zinc_binding_domain_zmout/1'>allosteric binding sites</scene> of the enzyme which exhibits cooperative binding. Four residues bind zinc with a high affinity even at 10<sup>-16</sup>M concentrations of Zinc in solution. Due to the tightness of Zinc binding, the complete enzyme has not yet been crystallized in its active conformation without the presence of Zinc metal inhibitor. When zinc is bound, DgcZ activity is limited<sup>[1]</sup>. Two Zinc binding sites are located on the CZB domain. | The <scene name='69/694239/Zb_domain_residues_19-90/5'>CZB Domain</scene>, residues 19-90, is responsible for regulating the function of DgcZ due to the presence of two zinc binding sites. The domain contains the <scene name='69/694239/Zinc_binding_domain_zmout/1'>allosteric binding sites</scene> of the enzyme which exhibits cooperative binding. Four residues bind zinc with a high affinity even at 10<sup>-16</sup>M concentrations of Zinc in solution. Due to the tightness of Zinc binding, the complete enzyme has not yet been crystallized in its active conformation without the presence of Zinc metal inhibitor. When zinc is bound, DgcZ activity is limited<sup>[1]</sup>. Two Zinc binding sites are located on the CZB domain. | ||
[[Image:Zinc binding site labels.jpg|250 px|left|thumb|Zn<sup>+2</sup> Coordination to amino acid residues on three of the four 𝝰 helices of DgcZ]] | [[Image:Zinc binding site labels.jpg|250 px|left|thumb|Zn<sup>+2</sup> Coordination to amino acid residues on three of the four 𝝰 helices of DgcZ]] | ||
| Line 36: | Line 36: | ||
Zahringer et al. mutated Cys52 to Ala through <span class="plainlinks">[https://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]</span>, resulting in a lack of coordination on α2. The cysteine residue is not essential for Zinc binding, as Zinc still coordinates to the three His residues with the Cys52Ala mutation, but α2 is free to move and expose the Zinc binding pocket. This exposure was found to lower the protein's affinity for zinc, as the mutation of cysteine to alanine increased the activity of the DgcZ. When not coordinated to zinc, the CZB domain presumably adopts a conformation that straightens the <scene name='69/694239/Czbd_with_helices_labeled/4'>α2 helix</scene>, shifting <scene name='69/694239/Hydrophobicity_int_residues/3'>hydrophobic residues</scene> on the α-helices into the center and the GGEEF domain into its productive conformation, increasing activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production. | Zahringer et al. mutated Cys52 to Ala through <span class="plainlinks">[https://en.wikipedia.org/wiki/Site-directed_mutagenesis site-directed mutagenesis]</span>, resulting in a lack of coordination on α2. The cysteine residue is not essential for Zinc binding, as Zinc still coordinates to the three His residues with the Cys52Ala mutation, but α2 is free to move and expose the Zinc binding pocket. This exposure was found to lower the protein's affinity for zinc, as the mutation of cysteine to alanine increased the activity of the DgcZ. When not coordinated to zinc, the CZB domain presumably adopts a conformation that straightens the <scene name='69/694239/Czbd_with_helices_labeled/4'>α2 helix</scene>, shifting <scene name='69/694239/Hydrophobicity_int_residues/3'>hydrophobic residues</scene> on the α-helices into the center and the GGEEF domain into its productive conformation, increasing activity of DgcZ. Activity increases without Zinc due to activation of poly-GlcNAc production and biofilm formation, and maximal cyclic di-GMP production. | ||
| - | + | ==Other Ligands== | |
c-di-GMP and GTP bind <scene name='69/694239/Flldgczwithc-di_and_allosteric/1'>allosterically</scene> although the function of this binding is unknown. Very weak product inhibition was observed when c-di-GMP bound allosterically but the inhibition was so weak, it is possible the c-di-GMP actually interacts with another as of yet unknown molecule at that site. | c-di-GMP and GTP bind <scene name='69/694239/Flldgczwithc-di_and_allosteric/1'>allosterically</scene> although the function of this binding is unknown. Very weak product inhibition was observed when c-di-GMP bound allosterically but the inhibition was so weak, it is possible the c-di-GMP actually interacts with another as of yet unknown molecule at that site. | ||
| - | == | + | ==See Also== |
| - | + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/3tvk 3tvk]</span> | |
| + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/3t9o 3t9o]</span> | ||
| + | <span class="plainlinks">[http://proteopedia.org/wiki/index.php/Diguanylate_cyclase Diguanylate cyclase]</span> | ||
== References == | == References == | ||
1. Zahringer, Franziska, Egidio Lancanna, Urs Jenal, Tilman Schirmer, and Alex Boehm. "Structure of E. Coli Zinc-Sensory Diguanylate Cyclase DgcZ." Cell Press: Structure 21 (2013): 1149-157.<span class="plainlinks">[http://ac.els-cdn.com/S0969212613001561/1-s2.0-S0969212613001561-main.pdf?_tid=912dc254-1635-11e7-9a5b-00000aacb35d&acdnat=1490980678_21f6bd7bbf72b51b6c6b3229a469afe5 doi: 10.1016/j.str.2013.04.026]</span> | 1. Zahringer, Franziska, Egidio Lancanna, Urs Jenal, Tilman Schirmer, and Alex Boehm. "Structure of E. Coli Zinc-Sensory Diguanylate Cyclase DgcZ." Cell Press: Structure 21 (2013): 1149-157.<span class="plainlinks">[http://ac.els-cdn.com/S0969212613001561/1-s2.0-S0969212613001561-main.pdf?_tid=912dc254-1635-11e7-9a5b-00000aacb35d&acdnat=1490980678_21f6bd7bbf72b51b6c6b3229a469afe5 doi: 10.1016/j.str.2013.04.026]</span> | ||
Revision as of 03:16, 21 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Diguanylate Cyclase DgcZ from Escherichia coli
| |||||||||||