We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1072
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
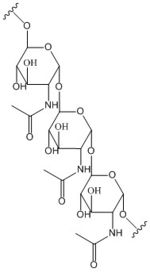
[[Image:Poly B-1, 6 GlcNAc.jpg |150 px|left|thumb|'''Figure 2: Poly-β-1,6-N-acetylglucosamine.''']] | [[Image:Poly B-1, 6 GlcNAc.jpg |150 px|left|thumb|'''Figure 2: Poly-β-1,6-N-acetylglucosamine.''']] | ||
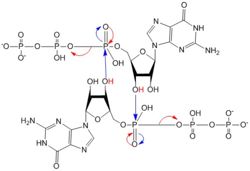
Diguanylate cyclases are a group of class 2 transferase enzymes that catalyze the production of cyclic dimeric-guanosine monophosphate (c-di-GMP, Figure 1), an important <span class="plainlinks">[https://en.wikipedia.org/wiki/Second_messenger_system second messenger]</span> for <span class="plainlinks">[https://en.wikipedia.org/wiki/Signal_transduction signal transduction]</span>.<sup>[1]</sup> Most commonly through phosphorylation or dephosphoylation events, signal transduction sends messages through cells to promote responses. <span class="plainlinks">[https://en.wikipedia.org/wiki/Escherichia_coli ''Escherechia coli'']</span>, a gram-negative bacterium often found in the intestines of mammals, uses diguanylate cyclase DgcZ in the synthesis of its <span class="plainlinks">[https://en.wikipedia.org/wiki/biofilm biofilm]</span>.<sup>[2]</sup> Enzyme DgcZ from ''E. coli'' acts as a catalyst to synthesize cyclic di-GMP from two substrate guanosine triphosphate (GTP) molecules to aid in communication of signals throughout the bacteria. C-di-GMP is a second messenger in the production of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc, Figure 2), a polysaccharide required for ''E. coli'' biofilm production <sup>[2]</sup>. This biofilm allows ''E. coli'' to adhere to extracellular surfaces. The complete enzyme is only successfully crystallized in its inactive conformation <sup>[3]</sup>. | Diguanylate cyclases are a group of class 2 transferase enzymes that catalyze the production of cyclic dimeric-guanosine monophosphate (c-di-GMP, Figure 1), an important <span class="plainlinks">[https://en.wikipedia.org/wiki/Second_messenger_system second messenger]</span> for <span class="plainlinks">[https://en.wikipedia.org/wiki/Signal_transduction signal transduction]</span>.<sup>[1]</sup> Most commonly through phosphorylation or dephosphoylation events, signal transduction sends messages through cells to promote responses. <span class="plainlinks">[https://en.wikipedia.org/wiki/Escherichia_coli ''Escherechia coli'']</span>, a gram-negative bacterium often found in the intestines of mammals, uses diguanylate cyclase DgcZ in the synthesis of its <span class="plainlinks">[https://en.wikipedia.org/wiki/biofilm biofilm]</span>.<sup>[2]</sup> Enzyme DgcZ from ''E. coli'' acts as a catalyst to synthesize cyclic di-GMP from two substrate guanosine triphosphate (GTP) molecules to aid in communication of signals throughout the bacteria. C-di-GMP is a second messenger in the production of poly-β-1,6-N-acetylglucosamine (poly-GlcNAc, Figure 2), a polysaccharide required for ''E. coli'' biofilm production <sup>[2]</sup>. This biofilm allows ''E. coli'' to adhere to extracellular surfaces. The complete enzyme is only successfully crystallized in its inactive conformation <sup>[3]</sup>. | ||
| - | [[Image:DgcZ_Conformation_change_1.png|250 px|right|thumb|'''Figure 3: Diagram of DgcZ.''' DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domain, which contains the Zinc binding sites<sup>[3]</sup>. Binding Zn<sup>+2</sup> inactivates the enzyme. The red vs blue coloring represents the symmetry of the enzyme.]] | + | [[Image:DgcZ_Conformation_change_1.png|250 px|right|thumb|'''Figure 3: Diagram of DgcZ.''' DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domain, which contains the Zinc binding sites<sup>[3]</sup>. Binding Zn<sup>+2</sup> inactivates the enzyme. The red vs blue coloring represents the symmetry of the enzyme.]]Image:C-di-GMP arrows.jpg |
== Structural Overview == | == Structural Overview == | ||
| Line 16: | Line 16: | ||
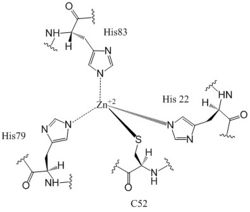
The <scene name='69/694239/Ggeef_domain_zmout_dgcz/2'>GGEEF domain</scene> of DgcZ is part of the GGDEF family of proteins that includes a conserved sequence, GG[DE][DE]F<sup>[6]</sup>.The GGEEF domain is a homodimer consisting of a central five-stranded β-sheet surrounded by five α-helices (Figure 4). The GGEEF domain contains two catalytic half sites that, when combined together in a productive conformation, form the entire <scene name='69/694239/Ggeef_domain_dgcz/6'>active site</scene>. Residues Gly-206, Gly-207, Glu-208, Glu-209, and Phe-210 form the active site. Each <scene name='69/694239/Ggeef_domain_half_site_dgcz/4'>half-site</scene> binds one GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to <scene name='69/694239/Gtp_guanine_bonds_asn_asp_dgcz/7'>Asn 173 and Asp 182</scene>. This scene depicts an inactive form of the protein because it was crystallized with zinc bound. The ribose of each guanosine triphosphate, and subsequent product c-di-GMP riboses, are held only loosely by the enzyme, while the phosphate groups are not bound to the active site at all<sup>[3]</sup>. | The <scene name='69/694239/Ggeef_domain_zmout_dgcz/2'>GGEEF domain</scene> of DgcZ is part of the GGDEF family of proteins that includes a conserved sequence, GG[DE][DE]F<sup>[6]</sup>.The GGEEF domain is a homodimer consisting of a central five-stranded β-sheet surrounded by five α-helices (Figure 4). The GGEEF domain contains two catalytic half sites that, when combined together in a productive conformation, form the entire <scene name='69/694239/Ggeef_domain_dgcz/6'>active site</scene>. Residues Gly-206, Gly-207, Glu-208, Glu-209, and Phe-210 form the active site. Each <scene name='69/694239/Ggeef_domain_half_site_dgcz/4'>half-site</scene> binds one GTP molecule. DgcZ binds the guanine base of GTP through hydrogen bonds to <scene name='69/694239/Gtp_guanine_bonds_asn_asp_dgcz/7'>Asn 173 and Asp 182</scene>. This scene depicts an inactive form of the protein because it was crystallized with zinc bound. The ribose of each guanosine triphosphate, and subsequent product c-di-GMP riboses, are held only loosely by the enzyme, while the phosphate groups are not bound to the active site at all<sup>[3]</sup>. | ||
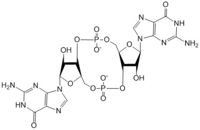
The alpha phosphate is available for attack by the 3 prime hydroxyl group on another GTP. A <scene name='69/694239/Gtp_magnesium_cofactors_dgcz/3'>Magnesium ion</scene> (Mg<sup>2+</sup>) stabilizes the negative charges on the phosphate groups. When in the productive conformation, each GTP is held in close proximity with the α-phosphate groups overlapping C3 of the ribose ring. This conformation allows the α-phospate of one GTP to react with the alcohol group attached to C3 of the ribose on the second GTP, resulting in a cyclization of the two molecules into c-di-GMP<sup>[3]</sup>. | The alpha phosphate is available for attack by the 3 prime hydroxyl group on another GTP. A <scene name='69/694239/Gtp_magnesium_cofactors_dgcz/3'>Magnesium ion</scene> (Mg<sup>2+</sup>) stabilizes the negative charges on the phosphate groups. When in the productive conformation, each GTP is held in close proximity with the α-phosphate groups overlapping C3 of the ribose ring. This conformation allows the α-phospate of one GTP to react with the alcohol group attached to C3 of the ribose on the second GTP, resulting in a cyclization of the two molecules into c-di-GMP<sup>[3]</sup>. | ||
| + | |||
===Mechanism of Action=== | ===Mechanism of Action=== | ||
| + | [[Image:C-di-GMP arrows.jpg|250 px|right|thumb|'''Figure 5: Mechanism of c-di-GMP synthesis.''']] | ||
Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap. | Diguanylate cyclases only function efficiently as dimers, to bind both GGDEF domains holding the substrates. The presence of Zinc disrupts the ability of the two domains to overlap. | ||
Revision as of 18:38, 21 April 2017
| This Sandbox is Reserved from 02/09/2015, through 05/31/2016 for use in the course "CH462: Biochemistry 2" taught by Geoffrey C. Hoops at the Butler University. This reservation includes Sandbox Reserved 1051 through Sandbox Reserved 1080. |
To get started:
More help: Help:Editing |
Diguanylate Cyclase DgcZ from Escherichia coli
| |||||||||||
![Figure 3: Diagram of DgcZ. DgcZ is shown in its active (left) and inactive (right) conformations. The boxes represent the GGEEF domains of the enzyme, while the cylinders represent the alpha helices of the CZB domain, which contains the Zinc binding sites[3]. Binding Zn+2 inactivates the enzyme. The red vs blue coloring represents the symmetry of the enzyme.](/wiki/images/thumb/d/df/DgcZ_Conformation_change_1.png/250px-DgcZ_Conformation_change_1.png)