We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1236
From Proteopedia
(Difference between revisions)
| Line 34: | Line 34: | ||
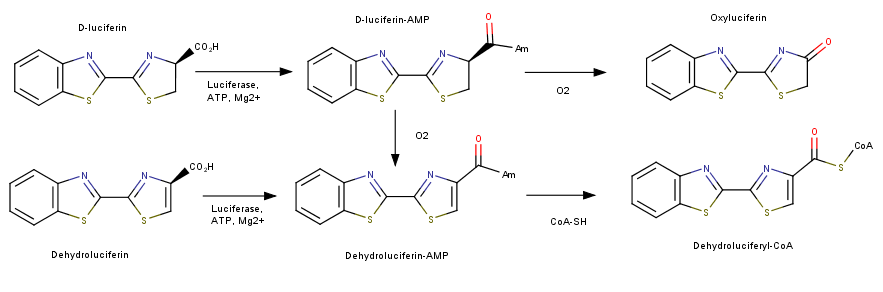
The attractive feature for the study and application of luciferase is its bioluminescent activity. The color of light produced has been found to vary depending on the organism that the protein is retrieved from. The fluorsence of light ranges through various wavelengths of light some of which include red, green and yellow light. This difference has a connection to the specificity of the active site of luciferase. Depending on the source of the protein different residues will produce a certain color of fluorescence. Variations in luciferase extracted from japanese fire flies has revealed this specificity. In high energy analogs of luciferase reveal the 286th residue is important in determine the color of light. When comparing green fluorescent japanese luciferin the active site contain an Ser286, while the red emitting variant has an Asp286. These findings indicate that slight variations in the active sight of luciferase can produce various effects on its bioluminencent activity. <ref>Goodsell, David . "Molecule of the Month: Luciferase." PDB-101: Luciferase. RSCB Protein Data Bank, June 2006. Web. 28 Apr. 2017.</ref> | The attractive feature for the study and application of luciferase is its bioluminescent activity. The color of light produced has been found to vary depending on the organism that the protein is retrieved from. The fluorsence of light ranges through various wavelengths of light some of which include red, green and yellow light. This difference has a connection to the specificity of the active site of luciferase. Depending on the source of the protein different residues will produce a certain color of fluorescence. Variations in luciferase extracted from japanese fire flies has revealed this specificity. In high energy analogs of luciferase reveal the 286th residue is important in determine the color of light. When comparing green fluorescent japanese luciferin the active site contain an Ser286, while the red emitting variant has an Asp286. These findings indicate that slight variations in the active sight of luciferase can produce various effects on its bioluminencent activity. <ref>Goodsell, David . "Molecule of the Month: Luciferase." PDB-101: Luciferase. RSCB Protein Data Bank, June 2006. Web. 28 Apr. 2017.</ref> | ||
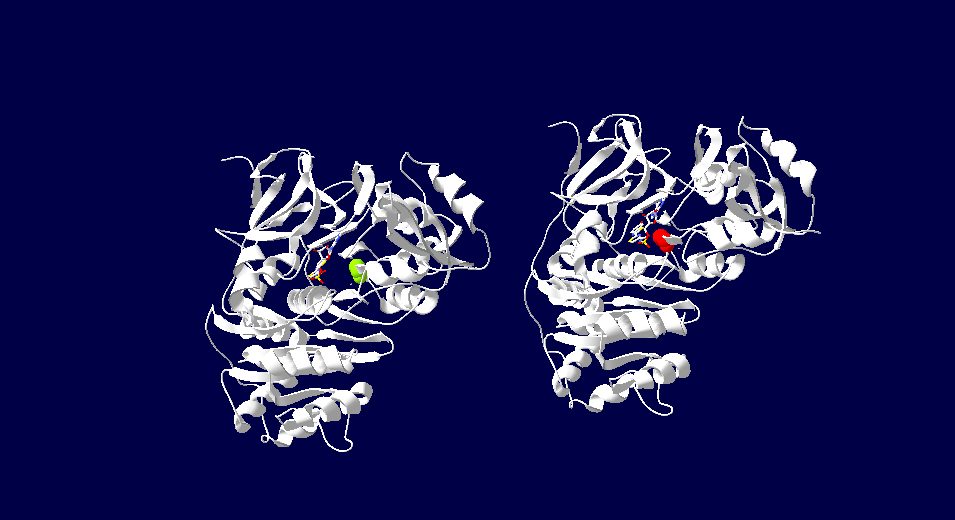
| - | [[Image: | + | [[Image:luciferasetwoview.png]] |
| - | + | This image depicts the two forms of Japanese firefly luciferase. The image on the left demonstrates green fluorescense due to its serine286. On the right the luciferase has red fluorescence due to the Asp286. The color of fluorescense is related to the specific amino acids in its active site. | |
| + | |||
==Inhibitors== | ==Inhibitors== | ||
Inhibition of luciferase can be mediated by several types of molecules. The presence of CoA has been found to have down regulate the bioluminescencent pathway of luciferase. Increased CoA favors the nonluminescent pathway which does not create a photon of light and instead produces Dehydroluciferyl-CoA. In addition to CoA inhibition of this enzyme has been seen with the exposure to diverse group of anesthetics. These molecules bind allosterically to luciferase and cause a conformation change in the protein. This unfolding alters the active site of the protein enough to not allow the bioluminescent catalysis to occur. Other than macro molecules, certain metal ions, specifically Ni and Co, have been seen to inhibit this reaction by replacing Magnesium in the active site. Magnesium is required for use of ATP and without it the reaction will not take place. Of the many inhibitors identified, lipoic acid demonstrates strong activity. Lipoic acid competes with luciferin for the active site enabling the binding of the substrate for the reaction. <ref>Leitão, João M. M., and Joaquim C G Esteves Da Silva. "Firefly luciferase inhibition." Journal of photochemistry and photobiology. (2010): 1-8. Research Gate. Web. 28 Apr. 2017.</ref> | Inhibition of luciferase can be mediated by several types of molecules. The presence of CoA has been found to have down regulate the bioluminescencent pathway of luciferase. Increased CoA favors the nonluminescent pathway which does not create a photon of light and instead produces Dehydroluciferyl-CoA. In addition to CoA inhibition of this enzyme has been seen with the exposure to diverse group of anesthetics. These molecules bind allosterically to luciferase and cause a conformation change in the protein. This unfolding alters the active site of the protein enough to not allow the bioluminescent catalysis to occur. Other than macro molecules, certain metal ions, specifically Ni and Co, have been seen to inhibit this reaction by replacing Magnesium in the active site. Magnesium is required for use of ATP and without it the reaction will not take place. Of the many inhibitors identified, lipoic acid demonstrates strong activity. Lipoic acid competes with luciferin for the active site enabling the binding of the substrate for the reaction. <ref>Leitão, João M. M., and Joaquim C G Esteves Da Silva. "Firefly luciferase inhibition." Journal of photochemistry and photobiology. (2010): 1-8. Research Gate. Web. 28 Apr. 2017.</ref> | ||
Revision as of 20:03, 1 May 2017
| This Sandbox is Reserved from Jan 17 through June 31, 2017 for use in the course Biochemistry II taught by Jason Telford at the Maryville University, St. Louis, USA. This reservation includes Sandbox Reserved 1225 through Sandbox Reserved 1244. |
To get started:
More help: Help:Editing |
Luciferase
| |||||||||||