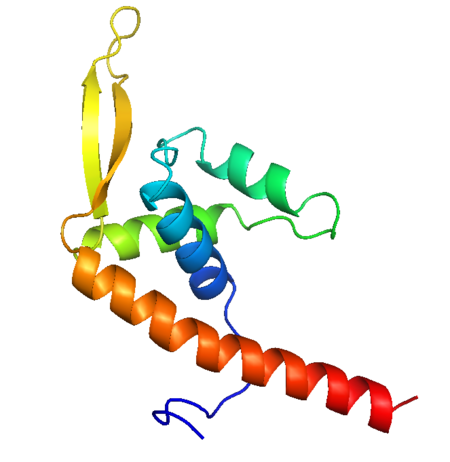
Replication Termination Protein
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
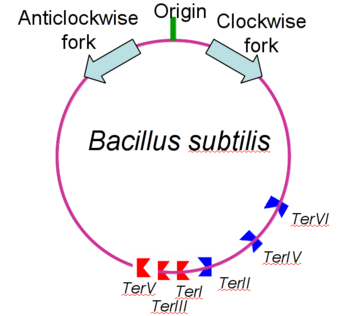
[[Image:yehhhh.png|350px|left|thumb| B. subtilis genome showing origin of replication, polar Ter sites and both replication forks.]] | [[Image:yehhhh.png|350px|left|thumb| B. subtilis genome showing origin of replication, polar Ter sites and both replication forks.]] | ||
| + | {{Clear}} | ||
Bacterial replication like that found in Bacillus subtilis consists of two replication forks that travel in opposite directions around the same circular strand of DNA. These replication forks begin at the origin of replication (OriC) and travel in clockwise and anticlockwise directions <ref> Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7 </ref>. To prevent the strand from being over replicated each strand needs to be terminated roughly opposite the OriC and then joined to form the complete duplicated strand of DNA. As such the Ter/RTP complexes have found to be polar in their action; that is that different Ter sites are capable of only blacking DNA replication from one direction only. Since Ter sites are found facing both forks of replication, and the combination of both are capable of stopping both strands of replication, they are often termed as replication traps. | Bacterial replication like that found in Bacillus subtilis consists of two replication forks that travel in opposite directions around the same circular strand of DNA. These replication forks begin at the origin of replication (OriC) and travel in clockwise and anticlockwise directions <ref> Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7 </ref>. To prevent the strand from being over replicated each strand needs to be terminated roughly opposite the OriC and then joined to form the complete duplicated strand of DNA. As such the Ter/RTP complexes have found to be polar in their action; that is that different Ter sites are capable of only blacking DNA replication from one direction only. Since Ter sites are found facing both forks of replication, and the combination of both are capable of stopping both strands of replication, they are often termed as replication traps. | ||
The arresting of replication is achieved by the binding of two dimers to each Ter site, with the two RTP binding sites referred to as site A and site B. Site B shows a higher affinity for RTP and binds first, with site A then being filled with another RTP dimer cooperatively. This differential affinity for RTP is cited as a possible reason for the observed polarity of the termination <ref> Duggin I.G. (2006) DNA Replication Fork Arrest by the Bacillus subtilis RTP–DNA Complex Involves a Mechanism that Is Independent of the Affinity of RTP–DNA Binding. Journal of Molecular Biology, 361:1-6 </ref>. | The arresting of replication is achieved by the binding of two dimers to each Ter site, with the two RTP binding sites referred to as site A and site B. Site B shows a higher affinity for RTP and binds first, with site A then being filled with another RTP dimer cooperatively. This differential affinity for RTP is cited as a possible reason for the observed polarity of the termination <ref> Duggin I.G. (2006) DNA Replication Fork Arrest by the Bacillus subtilis RTP–DNA Complex Involves a Mechanism that Is Independent of the Affinity of RTP–DNA Binding. Journal of Molecular Biology, 361:1-6 </ref>. | ||
Revision as of 10:09, 9 May 2017
| |||||||||||
3D structures of replication terminator protein
1bm9 – BsRTP – Bacillus subtilis
1j0r, 2dqr - BsRTP (mutant)
2dpd - BsRTP + DNA
1f4k, 2dpu, 2efw – BsRTP (mutant) + DNA
1ecr – Tus + DNA – Escherichia coli
References
- ↑ Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morkikawa K, (1996) Structure of a replication-terminator protein complexed with DNA. Nature, 383:598-603
- ↑ Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210
- ↑ Gautam A. et.al. (2001) A single domain of the replication termination protein of Bacillus subtilis is involved in arresting both DnaB helicase and RNA polymerase. Journal of Biological Chemistry, 276:23471-23479
- ↑ Noirot P (2007). "Replication of the Bacillus subtilis chromosome". In Graumann P. Bacillus: Cellular and Molecular Biology. Caister Academic Press. ISBN 978-1-904455-12-7
- ↑ Duggin I.G. (2006) DNA Replication Fork Arrest by the Bacillus subtilis RTP–DNA Complex Involves a Mechanism that Is Independent of the Affinity of RTP–DNA Binding. Journal of Molecular Biology, 361:1-6
- ↑ Vivian JP, Porter CJ, Wilce JA, Wilce MCJ, (2007) An asymmetric structure of the Bacillus subtilise Replication Terminator Protein in Complex with DNA. J. Mol. Bio, 370:481-491
- ↑ Vivian JP, Porter CJ, Wilce JA, Wilce MCJ, (2007) An asymmetric structure of the Bacillus subtilise Replication Terminator Protein in Complex with DNA. J. Mol. Bio, 370:481-491
- ↑ Bastia D. (1995) Crystal structure of the replication terminator protein from b. subtilis at 2.6 A. Cell 80: 651-660
- ↑ Wilce et. al. (2001) Structure of the RTP−DNA complex and the mechanism of polar replication fork arrest. Nature Structural Biology, 8:206-210
- ↑ Kaplan D.L., Bastia D. (2009). Mechanisms of polar arrest of a replication fork. Molecular Microbiology 72: 279-284
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Craig Mooney, Joel L. Sussman