This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
SecA
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
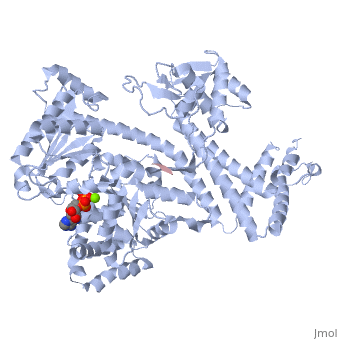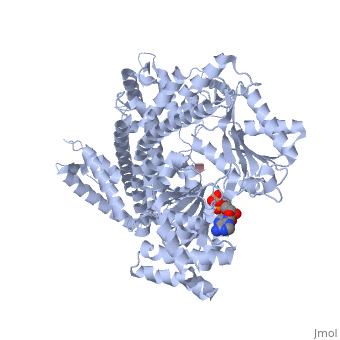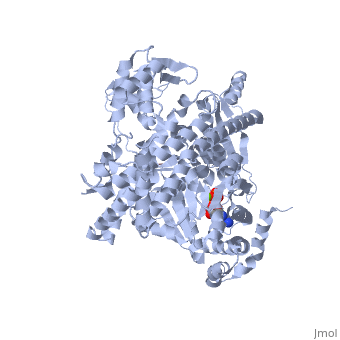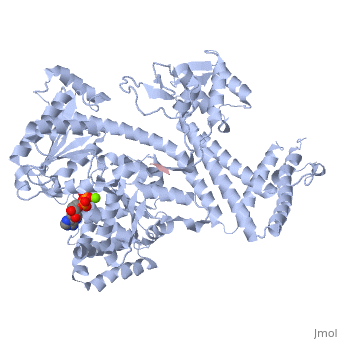
| - | <StructureSection load='3jv2' size='400' side='right' scene='' caption='Dimer of monomeric SecA complex with peptide (pink and yellow), ADP and Mg+2 ion (green), [[3jv2]]'> | + | <StructureSection load='3jv2' size='400' side='right' scene='' caption='Dimer of monomeric SecA complex with peptide (pink and yellow), ADP and Mg+2 ion (green), [[3jv2]]' pspeed='8'> |
=Introduction= | =Introduction= | ||
The [http://www.nature.com/nature/journal/v455/n7215/full/nature07335.html SecA] ATPase '''SecA''' drives the post-translational translocation of proteins through the SecY channel in the bacterial inner membrane. SecA is a dimer that can dissociate into monomers under certain conditions. Many bacterial proteins are transported post-translationally across the inner membrane by the Sec machinery, which consists of two essential components (1-4). One is the SecY complex, which forms a conserved heterotrimeric protein-conducting channel in the inner membrane.<ref name=journal1>PMID:15618215</ref> The other is SecA, a cytoplasmic ATPase, which "pushes" substrate polypeptide chains through the SecY channel.<ref name=journal1/> . For additional details see [[SecA PBD motions]]. | The [http://www.nature.com/nature/journal/v455/n7215/full/nature07335.html SecA] ATPase '''SecA''' drives the post-translational translocation of proteins through the SecY channel in the bacterial inner membrane. SecA is a dimer that can dissociate into monomers under certain conditions. Many bacterial proteins are transported post-translationally across the inner membrane by the Sec machinery, which consists of two essential components (1-4). One is the SecY complex, which forms a conserved heterotrimeric protein-conducting channel in the inner membrane.<ref name=journal1>PMID:15618215</ref> The other is SecA, a cytoplasmic ATPase, which "pushes" substrate polypeptide chains through the SecY channel.<ref name=journal1/> . For additional details see [[SecA PBD motions]]. | ||
| Line 7: | Line 7: | ||
Here we report crystal structures of SecA bound in an intermediate state of nucleotide hydrolysis to the SecY channel. The structures suggest mechanisms for how the channel is opened and prepared for the arrival of a translocation substrate, and how SecA moves polypeptides through the channel. | Here we report crystal structures of SecA bound in an intermediate state of nucleotide hydrolysis to the SecY channel. The structures suggest mechanisms for how the channel is opened and prepared for the arrival of a translocation substrate, and how SecA moves polypeptides through the channel. | ||
| - | Notable finding is that ADP binding to the high-affinity site stabilizes a compact conformation of SecA (ground state) that has low affinity for the SecYEG/membrane<ref name=journal1/>. Click the green link to view the active site of <scene name=' | + | Notable finding is that ADP binding to the high-affinity site stabilizes a compact conformation of SecA (ground state) that has low affinity for the SecYEG/membrane<ref name=journal1/>. Click the green link to view the active site of <scene name='38/382930/Cv/3'>ADP</scene> with surrounding amino acids (Water molecules shown as red spheres). This result suggests that following ATP hydrolysis, the ADP-bound SecA undergoes retraction from the translocon to complete one reaction cycle. However, the apo (nucleotide free) form of SecA can also exist in a compact conformation with low affinity for the translocon<ref name=journal1/>. Moreover, ADP release from SecA is stimulated by SecYEG/membrane, raising the question whether SecA retraction from the membrane occurs in the ADP-bound form, the apo form, or both<ref name=journal1/>. |
==Structure Determination Of SecA-SecY Complexes== | ==Structure Determination Of SecA-SecY Complexes== | ||
Revision as of 09:32, 23 May 2017
| |||||||||||
3D structures of SecA
23-May-2017
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 1.14 1.15 1.16 Or E, Boyd D, Gon S, Beckwith J, Rapoport T. The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem. 2005 Mar 11;280(10):9097-105. Epub 2004 Dec 23. PMID:15618215 doi:10.1074/jbc.M413947200
- ↑ 2.0 2.1 2.2 2.3 2.4 Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008 Oct 16;455(7215):936-43. PMID:18923516 doi:10.1038/nature07335
- ↑ Herbort M, Klein M, Manting EH, Driessen AJ, Freudl R. Temporal expression of the Bacillus subtilis secA gene, encoding a central component of the preprotein translocase. J Bacteriol. 1999 Jan;181(2):493-500. PMID:9882663
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Raffi Elmajian, Alexander Berchansky, Andrea Gorrell, David Canner