Sandbox Reserved 1053
From Proteopedia
| Line 3: | Line 3: | ||
== Background == | == Background == | ||
===Operon Overview=== | ===Operon Overview=== | ||
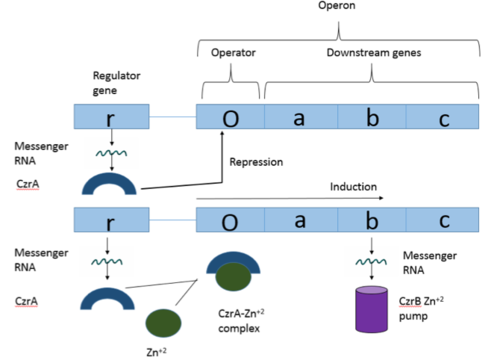
| - | [https://en.wikipedia.org/wiki/Operon Operons] are a critical genetic component of most prokaryotic cells. There are many different operons, responsible for the production of proteins with a wide range of functions. The most well-known and studied operons are the Lac and Trp operons, responsible for producing enzymes which metabolize lactose and tryptophan respectively. Despite many differences in each operon and the proteins that they encode, operons all function in the same general manner (Figure 1). Structurally, each operon contains a regulator, an operator, and one or more structural genes. The regulator gene codes for a protein responsible for managing the expression level of the structural genes. The operator contains the binding sequence for [https://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase] and is the site where transcription begins. Lastly, the structural genes code for proteins to be used elsewhere. The regulator protein (produced as a result of expression of the regulator gene) most often acts in a repressive manner, though this is not always the case. That is, the regulator protein will bind to the operator, inhibiting the binding and/or progression of RNA polymerase to the structural genes, thus inhibiting transcription of the genes into mRNA. If the regulator protein were always active, there could never be adequate expression of the structural genes, so there must be a way to inactive the regulator protein, thus enabling expression of the structural genes. This is usually achieved through the binding of an inhibitor to the regulator protein. Since regulator proteins are DNA binding proteins, often this inhibition is allosteric rather than competitive, that is the inhibitor is not something that mimics DNA and binds to the active site physically blocking DNA from binding. Rather, the inhibitor of the regulator binds to somewhere other than the active site of the protein, changing it in some way which decreases the proteins affinity or ability to bind DNA and repress transcription. | + | [https://en.wikipedia.org/wiki/Operon Operons] are a critical genetic component of most prokaryotic cells. There are many different operons, responsible for the production of proteins with a wide range of functions. The most well-known and studied operons are the [https://en.wikipedia.org/wiki/Lac_operon Lac] and [https://en.wikipedia.org/wiki/Trp_operon Trp] operons, responsible for producing enzymes which metabolize lactose and tryptophan respectively. Despite many differences in each operon and the proteins that they encode, operons all function in the same general manner (Figure 1). Structurally, each operon contains a [https://en.wikipedia.org/wiki/Regulator_gene regulator], an [https://en.wikipedia.org/wiki/Operator_(biology) operator], and one or more [https://en.wikipedia.org/wiki/Structural_gene structural genes]. The regulator gene codes for a protein responsible for managing the expression level of the structural genes. The operator contains the binding sequence for [https://en.wikipedia.org/wiki/RNA_polymerase RNA polymerase] and is the site where [https://en.wikipedia.org/wiki/Transcription_(biology) transcription] begins. Lastly, the structural genes code for proteins to be used elsewhere. The regulator protein (produced as a result of expression of the regulator gene) most often acts in a repressive manner, though this is not always the case. That is, the regulator protein will bind to the operator, inhibiting the binding and/or progression of RNA polymerase to the structural genes, thus inhibiting transcription of the genes into mRNA. If the regulator protein were always active, there could never be adequate expression of the structural genes, so there must be a way to inactive the regulator protein, thus enabling expression of the structural genes. This is usually achieved through the binding of an inhibitor to the regulator protein. Since regulator proteins are DNA binding proteins, often this inhibition is [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric] rather than competitive, that is the inhibitor is not something that mimics DNA and binds to the active site physically blocking DNA from binding. Rather, the inhibitor of the regulator binds to somewhere other than the active site of the protein, changing it in some way which decreases the proteins affinity or ability to bind DNA and repress transcription. |
[[Image:Operon.png|500px|thumb|center|Figure 1: Overview of Operon Structure]] | [[Image:Operon.png|500px|thumb|center|Figure 1: Overview of Operon Structure]] | ||
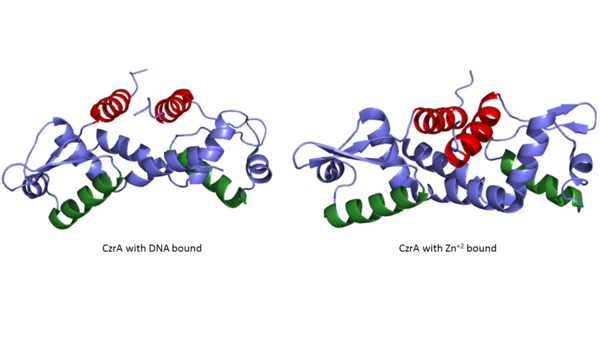
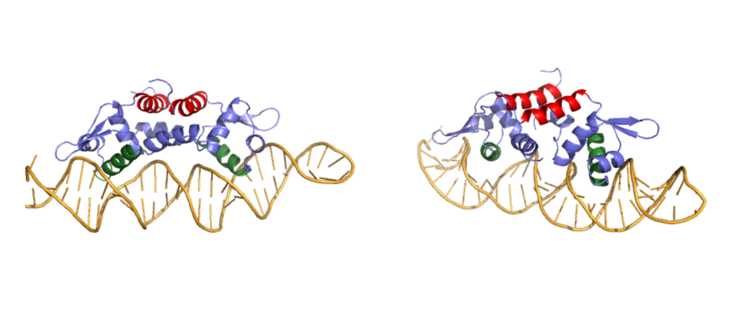
<StructureSection load='CzrAwithDNA.pdb' size='350' frame='true' align='left' caption='Czr A dimer bound to DNA' scene='Insert optional scene name here' /> | <StructureSection load='CzrAwithDNA.pdb' size='350' frame='true' align='left' caption='Czr A dimer bound to DNA' scene='Insert optional scene name here' /> | ||
| Line 36: | Line 36: | ||
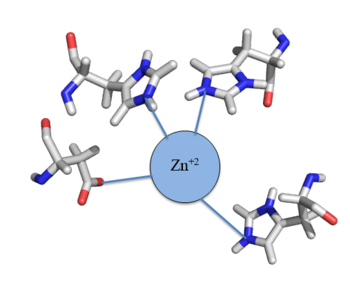
[[Image:Zinc tetrahedral complex.PNG|350px|thumb|center| Figure 4: Zn<sup>+2</sup> tetrahedral binding complex]] | [[Image:Zinc tetrahedral complex.PNG|350px|thumb|center| Figure 4: Zn<sup>+2</sup> tetrahedral binding complex]] | ||
| - | Zinc<sup>+2</sup> binding is driven by a large [https://en.wikipedia.org/wiki/Entropy entropic] gain <ref>DOI:10.1021/ja906131b</ref>. Water molecules around the metal ion and CzrA protein are displaced, and gain greater freedom. This gain in entropy allows Zn<sup>+2</sup> to bind to CzrA with reasonable affinity and speed in vivo. The zinc<sup>+2</sup> ion forms a tetrahedral complex with the four residues (Figure | + | Zinc<sup>+2</sup> binding is driven by a large [https://en.wikipedia.org/wiki/Entropy entropic] gain <ref>DOI:10.1021/ja906131b</ref>. Water molecules around the metal ion and CzrA protein are displaced, and gain greater freedom. This gain in entropy allows Zn<sup>+2</sup> to bind to CzrA with reasonable affinity and speed in vivo. The zinc<sup>+2</sup> ion forms a tetrahedral complex with the four residues (Figure 4). This allows other metal ions to act as allosteric inhibitors to CzrA. Any metal that may form a tetrahedral complex will have some affinity for CzrA, assuming it is not too large to fit into the pocket. However, the metal binding pocket of CzrA has been optimized to bind Zn<sup>+2</sup> with the highest affinity. As CzrA is a transcriptional repressor, binding of Zn<sup>+2</sup> to the dimer will activate the czr operon. Zn<sup>+2</sup> is preferred as CzrB opens a Zn<sup>+2</sup> channel, allowing the excess zinc ions to export the cell. |
</StructureSection> | </StructureSection> | ||
Revision as of 20:30, 15 August 2017
CzrA: A Zinc Dependent Transcriptional Regulator
Background
Operon Overview
Operons are a critical genetic component of most prokaryotic cells. There are many different operons, responsible for the production of proteins with a wide range of functions. The most well-known and studied operons are the Lac and Trp operons, responsible for producing enzymes which metabolize lactose and tryptophan respectively. Despite many differences in each operon and the proteins that they encode, operons all function in the same general manner (Figure 1). Structurally, each operon contains a regulator, an operator, and one or more structural genes. The regulator gene codes for a protein responsible for managing the expression level of the structural genes. The operator contains the binding sequence for RNA polymerase and is the site where transcription begins. Lastly, the structural genes code for proteins to be used elsewhere. The regulator protein (produced as a result of expression of the regulator gene) most often acts in a repressive manner, though this is not always the case. That is, the regulator protein will bind to the operator, inhibiting the binding and/or progression of RNA polymerase to the structural genes, thus inhibiting transcription of the genes into mRNA. If the regulator protein were always active, there could never be adequate expression of the structural genes, so there must be a way to inactive the regulator protein, thus enabling expression of the structural genes. This is usually achieved through the binding of an inhibitor to the regulator protein. Since regulator proteins are DNA binding proteins, often this inhibition is allosteric rather than competitive, that is the inhibitor is not something that mimics DNA and binds to the active site physically blocking DNA from binding. Rather, the inhibitor of the regulator binds to somewhere other than the active site of the protein, changing it in some way which decreases the proteins affinity or ability to bind DNA and repress transcription.
|
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Arunkumar A., Campanello G., Giedroc D. (2009). Solution Structure of a paradigm ArsR family zinc sensor in the DNA-bound state. PNAS 106:43 18177-18182.
- ↑ MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. PMID:16959962 doi:http://dx.doi.org/10.1128/MMBR.00015-06
- ↑ Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.
- ↑ Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009 Dec 16;131(49):17860-70. doi: 10.1021/ja906131b. PMID:19995076 doi:http://dx.doi.org/10.1021/ja906131b