Sarcosine oxidase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
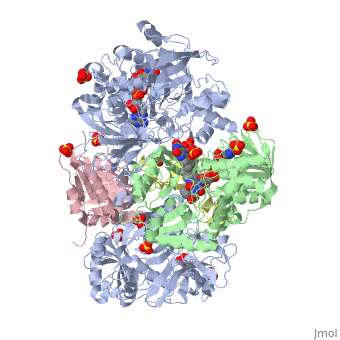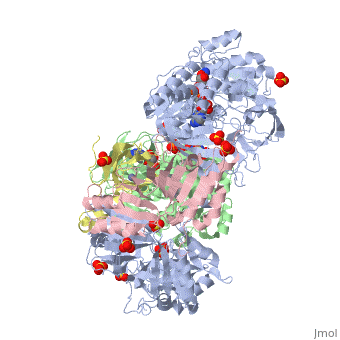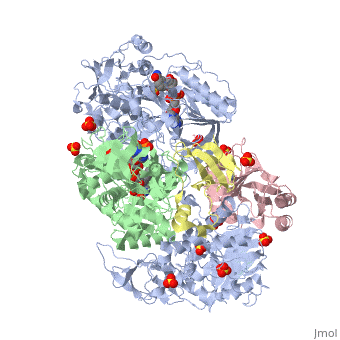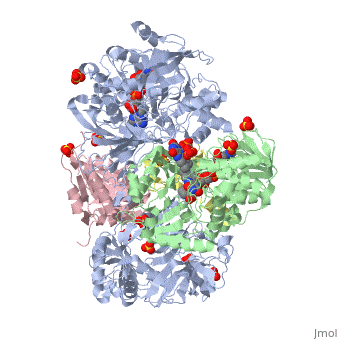
| - | + | <StructureSection load='3ad8' size='350' side='right' scene='' caption='Heterotetramer of sarcosine oxidase containing FAD, FMN and NAD showing α subunit (grey), β subunit (green), γ subunit (pink) and δ subunit (yellow). Complex with pyrrole-2-carboxylate, sulfate and Zn+2 ions (grey) [[3ad8]]'> | |
| - | <StructureSection load='3ad8' size=' | + | |
'''Sarcosine oxidase''' (SOX) catalyzes the demethylation of sarcosine+O2+H4-tetrahydrofolate to produce glycine, hydrogen peroxide and 5, 10-methylene-tetrahydrofolate. Sarcosine metabolism can be the source of carbon and energy for many microorganisms. SOXs are found as monomers, heterodirmers and heterotetramers<ref>PMID:24185971</ref>. Monomeric SOX uses FAD as cofactor. Heterotetrameric SOX uses FAD and FMN as cofactors. | '''Sarcosine oxidase''' (SOX) catalyzes the demethylation of sarcosine+O2+H4-tetrahydrofolate to produce glycine, hydrogen peroxide and 5, 10-methylene-tetrahydrofolate. Sarcosine metabolism can be the source of carbon and energy for many microorganisms. SOXs are found as monomers, heterodirmers and heterotetramers<ref>PMID:24185971</ref>. Monomeric SOX uses FAD as cofactor. Heterotetrameric SOX uses FAD and FMN as cofactors. | ||
Revision as of 17:54, 17 September 2018
| |||||||||||
3D structures of sarcosine oxidase
Updated on 17-September-2018
References
- ↑ Suzuki H. Sarcosine oxidase: structure, function, and the application to creatinine determination. Amino Acids. 1994 Feb;7(1):27-43. doi: 10.1007/BF00808444. PMID:24185971 doi:http://dx.doi.org/10.1007/BF00808444