We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Transposase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
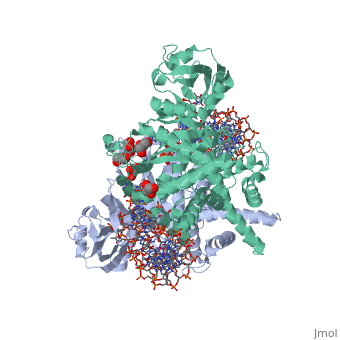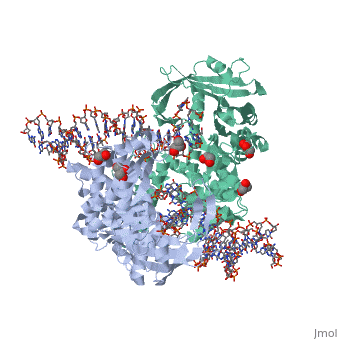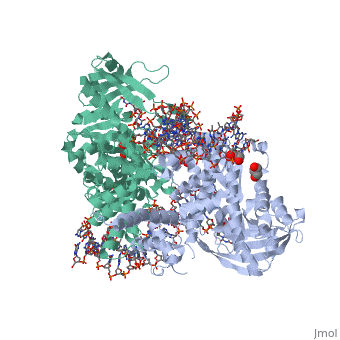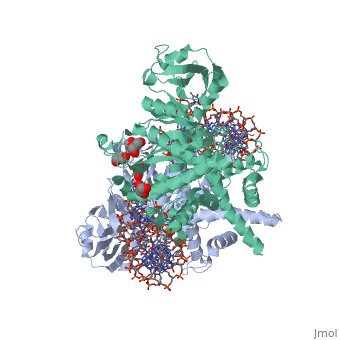
| - | <StructureSection load='3ecp' size=' | + | <StructureSection load='3ecp' size='350' side='right' caption='Structure of Tn5 transposase complex with DNA and glycerol (PDB code [[3ecp]]).' scene=''> |
__TOC__ | __TOC__ | ||
== Function == | == Function == | ||
| Line 27: | Line 27: | ||
**[[1bcm]], [[1bco]] – MuMuA core domain <br /> | **[[1bcm]], [[1bco]] – MuMuA core domain <br /> | ||
**[[2ezh]], [[2ezi]] – MuMuA Iγ subdomain - NMR<br /> | **[[2ezh]], [[2ezi]] – MuMuA Iγ subdomain - NMR<br /> | ||
| - | **[[2ezk]] – MuMuA Iβ subdomain - NMR<br /> | + | **[[2ezk]], [[2ezl]] – MuMuA Iβ subdomain - NMR<br /> |
**[[4fcy]] – MuMuA + DNA <br /> | **[[4fcy]] – MuMuA + DNA <br /> | ||
| Line 33: | Line 33: | ||
**[[2bw3]] – fTra (mutant) - fly<br /> | **[[2bw3]] – fTra (mutant) - fly<br /> | ||
| - | **[[4d1q]] – fTra (mutant) + DNA <br /> | + | **[[4d1q]], [[6dx0]], [[6dww]], [[6dwy]], [[6dwz]] – fTra (mutant) + DNA <br /> |
*Dra2 transposase | *Dra2 transposase | ||
| Line 69: | Line 69: | ||
**[[5cr4]] – SB catalytic domain – synthetic<br /> | **[[5cr4]] – SB catalytic domain – synthetic<br /> | ||
| + | **[[2m8e]] – SB PAI domain – synthetic - NMR<br /> | ||
**[[5unk]] – RED domain – ''Oncorhynchus mykiss'' - NMR<br /> | **[[5unk]] – RED domain – ''Oncorhynchus mykiss'' - NMR<br /> | ||
Revision as of 22:59, 26 September 2018
| |||||||||||
3D structures of transposase
Updated on 26-September-2018
References
- ↑ Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009 Jun;6(6):415-22. doi: 10.1038/nmeth.1332. PMID:19478801 doi:http://dx.doi.org/10.1038/nmeth.1332
- ↑ Rice P, Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995 Jul 28;82(2):209-20. PMID:7628012
- ↑ Park JM, Evertts AG, Levin HL. The Hermes transposon of Musca domestica and its use as a mutagen of Schizosaccharomyces pombe. Methods. 2009 Nov;49(3):243-7. doi: 10.1016/j.ymeth.2009.05.004. Epub 2009 May, 18. PMID:19450689 doi:http://dx.doi.org/10.1016/j.ymeth.2009.05.004
- ↑ Reznikoff WS. Transposon Tn5. Annu Rev Genet. 2008;42:269-86. PMID:18680433 doi:http://dx.doi.org/10.1146/annurev.genet.42.110807.091656
- ↑ Craig NL. Tn7: a target site-specific transposon. Mol Microbiol. 1991 Nov;5(11):2569-73. PMID:1664019
- ↑ Wang W, Swevers L, Iatrou K. Mariner (Mos1) transposase and genomic integration of foreign gene sequences in Bombyx mori cells. Insect Mol Biol. 2000 Apr;9(2):145-55. PMID:10762422
- ↑ Colloms SD, van Luenen HG, Plasterk RH. DNA binding activities of the Caenorhabditis elegans Tc3 transposase. Nucleic Acids Res. 1994 Dec 25;22(25):5548-54. PMID:7838706
- ↑ Ivics Z, Kaufman CD, Zayed H, Miskey C, Walisko O, Izsvak Z. The Sleeping Beauty transposable element: evolution, regulation and genetic applications. Curr Issues Mol Biol. 2004 Jan;6(1):43-55. PMID:14632258
- ↑ Klenchin VA, Czyz A, Goryshin IY, Gradman R, Lovell S, Rayment I, Reznikoff WS. Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif. Nucleic Acids Res. 2008 Oct;36(18):5855-62. Epub 2008 Sep 12. PMID:18790806 doi:10.1093/nar/gkn577