We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Monooxygenase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
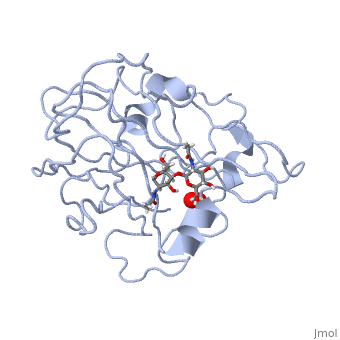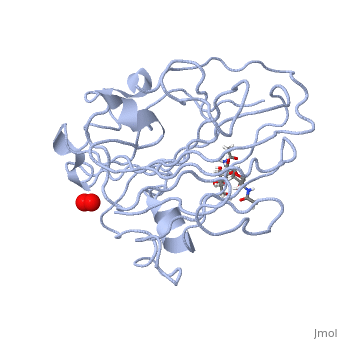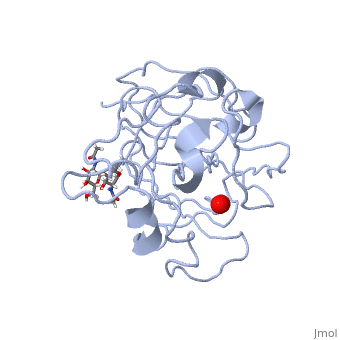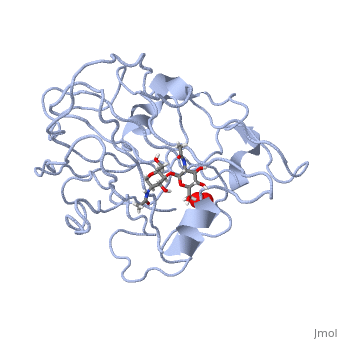
| - | <StructureSection load='5tkh' size='350' side='right' scene='' caption=' | + | <StructureSection load='5tkh' size='350' side='right' scene='' caption='Glycosylated lytic polysaccharide monooxygenase complex with Cu(II) (orange) and peroxide (PDB code [[5tkh]])' > |
== Function == | == Function == | ||
Revision as of 15:12, 20 October 2017
| |||||||||||
3D structures of monooxygenase
Updated on 20-October-2017
Methane monooxygenase See Methane monooxygenase
Camphor 5-monooxygenase See Cytochrome P450
Luciferin 4-monooxygenase and Alkanal monooxygenase See Luciferase
References
- ↑ Rudzka K, Moreno DM, Eipper B, Mains R, Estrin DA, Amzel LM. Coordination of peroxide to the Cu(M) center of peptidylglycine alpha-hydroxylating monooxygenase (PHM): structural and computational study. J Biol Inorg Chem. 2012 Dec 18. PMID:23247335 doi:10.1007/s00775-012-0967-z
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky