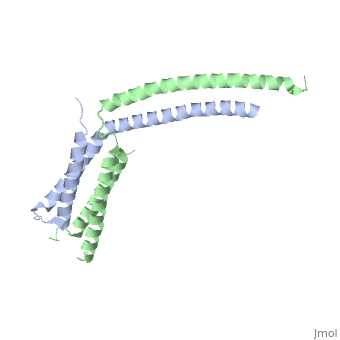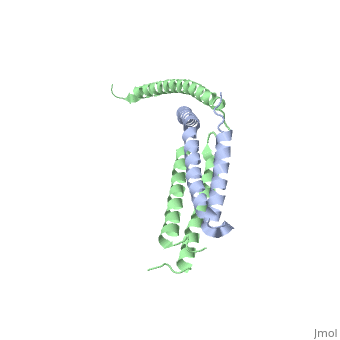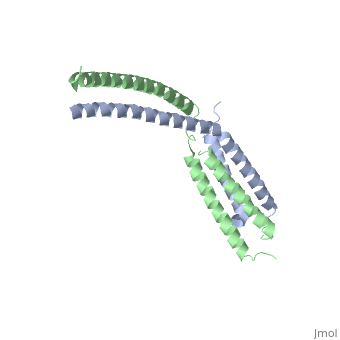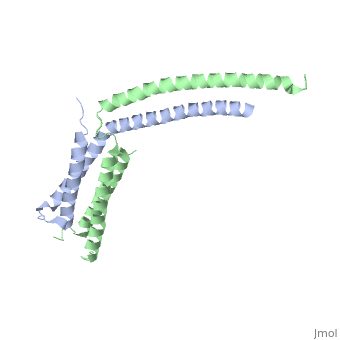
Mediator
From Proteopedia
| Line 2: | Line 2: | ||
== Mediator == | == Mediator == | ||
| - | The '''mediator''', found in the yeast ''S. cerevisiae'', connects repressors and activators bound to regulatory DNA with RNA polymerase II (Pol II). As a result, the mediator is an important regulator of eukaryotic transcription.<ref>PMID: 21725323 </ref> Also, mediator is currently being studied because of its ability and role to regulate gene expression.<ref>PMID: 20299225 </ref> Mediator is in the form of three main modules, which are the tail, arm, and head. The <scene name='Sandbox_Reserved_387/Head_module/1'>head module</scene> of the mediator is split into three domains that go through significant conformational changes, and it interacts with the Rpb7 and Rpb4 subunits of Pol II and TATA binding protein subunit of transcription factor TFIID.<ref name="nature">PMID: 21725323 </ref> | + | The '''mediator'''or '''mediator of RNA polymerase II transcription subunit''', found in the yeast ''S. cerevisiae'', connects repressors and activators bound to regulatory DNA with RNA polymerase II (Pol II). As a result, the mediator is an important regulator of eukaryotic transcription.<ref>PMID: 21725323 </ref> Also, mediator is currently being studied because of its ability and role to regulate gene expression.<ref>PMID: 20299225 </ref> Mediator is in the form of three main modules, which are the tail, arm, and head. The <scene name='Sandbox_Reserved_387/Head_module/1'>head module</scene> of the mediator is split into three domains that go through significant conformational changes, and it interacts with the Rpb7 and Rpb4 subunits of Pol II and TATA binding protein subunit of transcription factor TFIID.<ref name="nature">PMID: 21725323 </ref> |
| Line 13: | Line 13: | ||
==3D structures of mediator== | ==3D structures of mediator== | ||
| - | [[3rj1]] – | + | [[3rj1]] – yMed6 + Med8 + Med11 + Med17 + Med18 + Med20 + Med22 - yeast<br /> |
| - | [[1yke]], [[1ykh]] – | + | [[5u0s]] – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med27 + Med31 + RPB1 + RPB2 + RPB3 + RPB4 + RPB5 + RPB6 + RPB7 + RPB8 + RPB9 + RPB10 + RPB11 + RPB12 – fission yeast – Cryo EM<br /> |
| - | [[3fbi]], [[3fbn]] – | + | [[5u0s]] – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med27 + Med31 – Cryo EM<br /> |
| - | [[3c0t]] - | + | [[5n9j]] – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med31 <br /> |
| - | [[2hzs]] – | + | [[5sva]] – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med11 + Med17 + Med18 + Med20 + Med21 + Med22 + Med31 + pre-initiation complex – Cryo EM<br /> |
| - | [[3r84]] - | + | [[4v1o]] – yMed8 + Med11 + Med18 + Med20 + Med22 + pre-initiation complex – Cryo EM<br /> |
| - | [[2lpb]] - | + | [[1yke]], [[1ykh]] – yMed7 + Med21 <br /> |
| - | [[2hzm]] – | + | [[3fbi]], [[3fbn]] – yMed7 N terminal + Med31<br /> |
| - | [[2l23]], [[2l6u]], [[2ky6]], [[2xnf]] – | + | [[3c0t]] - fyMed8 C terminal + Med18<br /> |
| - | + | [[2hzs]] – yMed8 C terminal + Med18 + Med20<br /> | |
| + | [[3r84]] - yMed11 + Med22 (mutant)<br /> | ||
| + | [[6aly]] – yMed 15 ABD2 residues<br /> | ||
| + | [[2lpb]] - yMed15 + general control protein GCN4<br /> | ||
| + | [[2hzm]] – yMed18 + Med20<br /> | ||
| + | [[2l23]], [[2l6u]], [[2ky6]], [[2xnf]] – hMed25 activator-interacting domain - human - NMR<br /> | ||
| + | [[5odd]] – hMed26 N terminal<br /> | ||
Revision as of 09:08, 6 August 2018
| |||||||||||
3D structures of mediator
3rj1 – yMed6 + Med8 + Med11 + Med17 + Med18 + Med20 + Med22 - yeast
5u0s – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med27 + Med31 + RPB1 + RPB2 + RPB3 + RPB4 + RPB5 + RPB6 + RPB7 + RPB8 + RPB9 + RPB10 + RPB11 + RPB12 – fission yeast – Cryo EM
5u0s – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med27 + Med31 – Cryo EM
5n9j – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med10 + Med11 + Med14 + Med17 + Med18 + Med19 + Med20 + Med21 + Med22 + Med31
5sva – fyMed4 + Med6 + Med7 + Med8 + Med9 + Med11 + Med17 + Med18 + Med20 + Med21 + Med22 + Med31 + pre-initiation complex – Cryo EM
4v1o – yMed8 + Med11 + Med18 + Med20 + Med22 + pre-initiation complex – Cryo EM
1yke, 1ykh – yMed7 + Med21
3fbi, 3fbn – yMed7 N terminal + Med31
3c0t - fyMed8 C terminal + Med18
2hzs – yMed8 C terminal + Med18 + Med20
3r84 - yMed11 + Med22 (mutant)
6aly – yMed 15 ABD2 residues
2lpb - yMed15 + general control protein GCN4
2hzm – yMed18 + Med20
2l23, 2l6u, 2ky6, 2xnf – hMed25 activator-interacting domain - human - NMR
5odd – hMed26 N terminal
References
- ↑ Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Architecture of the Mediator head module. Nature. 2011 Jul 3;475(7355):240-3. doi: 10.1038/nature10162. PMID:21725323 doi:10.1038/nature10162
- ↑ Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010 Jun;35(6):315-22. Epub 2010 Mar 17. PMID:20299225 doi:10.1016/j.tibs.2010.02.004
- ↑ 3.0 3.1 Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Architecture of the Mediator head module. Nature. 2011 Jul 3;475(7355):240-3. doi: 10.1038/nature10162. PMID:21725323 doi:10.1038/nature10162
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Joel L. Sussman, Bradley Altier, Alexander Berchansky