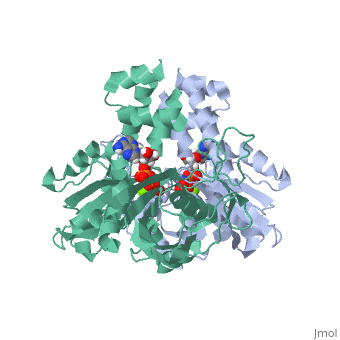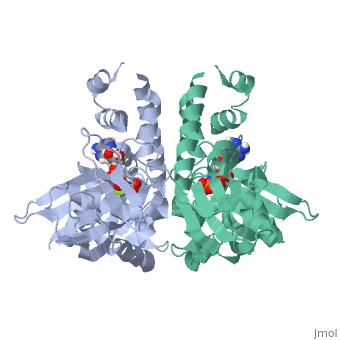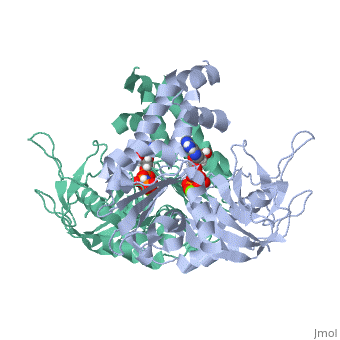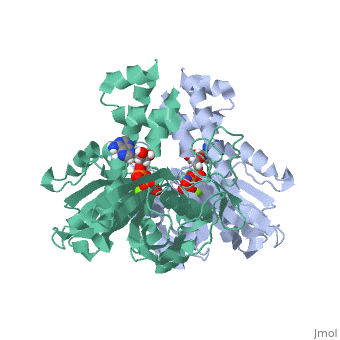This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Plasmid segregation protein ParM
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Structural highlights == | == Structural highlights == | ||
ParM polymerizes into double helical protofilaments with repeats similar to F-actin. <scene name='71/715423/Cv/2'>AMPPNP binding site</scene>. | ParM polymerizes into double helical protofilaments with repeats similar to F-actin. <scene name='71/715423/Cv/2'>AMPPNP binding site</scene>. | ||
| - | *<scene name='71/715423/Cv/3'>Mg coordination site</scene>. Water molecules shown as red spheres. | + | *<scene name='71/715423/Cv/3'>Mg coordination site</scene>. Water molecules are shown as red spheres. |
Revision as of 12:48, 7 August 2019
| |||||||||||
3D structures of plasmid segregation protein ParM
Updated on 07-August-2019
1mwk – ParM – Escherichias coli
1mwm – ParM + ADP
2zgy – ParM + GDP
2zgz – ParM + GMPPNP
4a61 – ParM + AMPPNP
5aey – ParM + AMPPNP - CryoEM
4a62 – ParM + ParR C terminus
2qu4, 2ghc, 3iku, 3iky, 4a6j, 5ai7 – ParM – CryoEM
2zhc – ParM + ADP - CryoEM
References
- ↑ Salje J, Gayathri P, Lowe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010 Oct;8(10):683-92. doi: 10.1038/nrmicro2425. PMID:20844556 doi:http://dx.doi.org/10.1038/nrmicro2425