Z-DNA
From Proteopedia
| Line 1: | Line 1: | ||
| - | <StructureSection load='Z-DNA.pdb' size=' | + | <StructureSection load='Z-DNA.pdb' size='350' side='right' scene='Z-DNA/Z-dna_new/1' caption=''> |
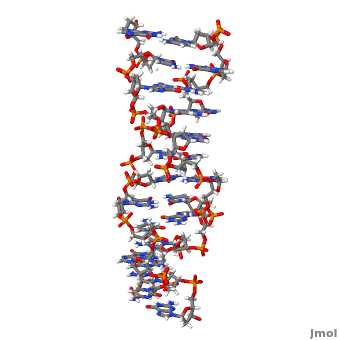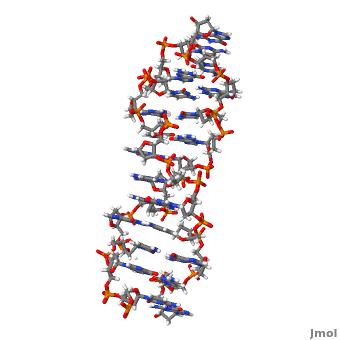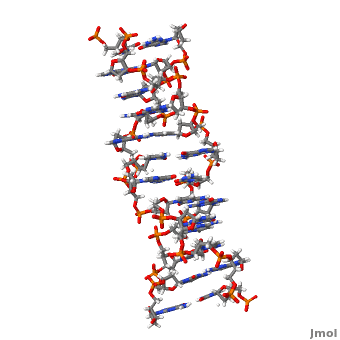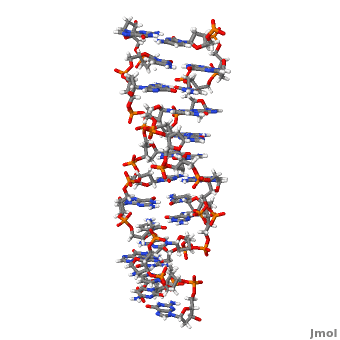
'''Z-DNA''' <scene name='Z-DNA/Z-dna_new/1'>(default scene)</scene> is a form of DNA that has a different structure from the more common <scene name='Sandbox_Z-DNA/Bdna/3'>B-DNA</scene> form.It is a left-handed double helix wherein the sugar-phosphate backbone has a zigzag pattern due to the alternate stacking of bases in [http://proteopedia.org/wiki/index.php/Syn_and_anti_nucleosides anti-conformation and syn conformation]. In Z-DNA only a minor groove is present and the major groove is absent. The residues that allow sequence-specific recognition of Z-DNA are present on the convex outer surface.<ref name = 'Rich'> PMID:12838348</ref> This DNA form is thought to play a role in the regulation of gene expression, DNA processing events and/or genetic instability.<ref name = 'Wang'>PMID:17485386</ref> | '''Z-DNA''' <scene name='Z-DNA/Z-dna_new/1'>(default scene)</scene> is a form of DNA that has a different structure from the more common <scene name='Sandbox_Z-DNA/Bdna/3'>B-DNA</scene> form.It is a left-handed double helix wherein the sugar-phosphate backbone has a zigzag pattern due to the alternate stacking of bases in [http://proteopedia.org/wiki/index.php/Syn_and_anti_nucleosides anti-conformation and syn conformation]. In Z-DNA only a minor groove is present and the major groove is absent. The residues that allow sequence-specific recognition of Z-DNA are present on the convex outer surface.<ref name = 'Rich'> PMID:12838348</ref> This DNA form is thought to play a role in the regulation of gene expression, DNA processing events and/or genetic instability.<ref name = 'Wang'>PMID:17485386</ref> | ||
Current revision
| |||||||||||
Movie Depicting ADAR1 binding to Z-DNA
Comparison of the three helices and helical parameters of DNA
Sources[7]
|
|
|
| Parameter | A-DNA | B-DNA | Z-DNA |
|---|---|---|---|
| Helix sense | right-handed | right-handed | left-handed |
| Residues per turn | 11 | 10.5 | 12 |
| Axial rise [Å] | 2.55 | 3.4 | 3.7 |
| Helix pitch(°) | 28 | 34 | 45 |
| Base pair tilt(°) | 20 | −6 | 7 |
| Rotation per residue (°) | 33 | 36 | -30 |
| Diameter of helix [Å] | 23 | 20 | 18 |
| Glycosidic bond configuration dA,dT,dC dG | anti anti | anti anti | anti syn |
| Sugar pucker dA,dT,dC dG | C3'-endo C3'-endo | C2'-endo C2'-endo | C2'-endo C3'-endo |
| Intrastrand phosphate-phosphate distance [Å] dA,dT,dC dG | 5.9 5.9 | 7.0 7.0 | 7.0 5.9 |
| Sources:[8][9][10] | |||
3D structures of Z-DNA
Updated on 09-October-2018
1woe, 3qba, 1i0t, 6aqt, 6aqv, 6aqw, 6aqx, 2ie1, 1da1, 1ick, 313d, 314d, 390d, 391d, 392d, 3p4j, 3qba, 3wbo, 400d, 4fs5, 4fs6, 4hif, 4hig, 2hto, 2htt, 4r15, 4xsn, 6bst – ZDNA hexamer CGCGCG
1v9g – ZDNA hexamer CGCGCG - Neutron
1tne – ZDNA hexamer CGCGCG - NMR
2obz – ZDNA hexamer CGCGUG
362d – ZDNA hexamer TGCGCA
1qbj – ZDNA hexamer CGCGCG + hADAR1 Zα domain - human
2heo - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zα domain – mouse
1sfu - ZDNA hexamer CGCGCG + Pox virus Z-DNA-binding protein Zα domain
4wcg - ZDNA hexamer CGCGCG + Herpes virus 3 Z-DNA-binding protein Zα domain
1j75 - ZDNA hexamer CGCGCG + DLM-1 Z-DNA-binding protein Zα domain
3f21 - ZDNA hexamer CACGTG + double-stranded RNA-specific adenosine deaminase Z-DNA-binding protein Zα domain
3f21 - ZDNA hexamer CGTACG + double-stranded RNA-specific adenosine deaminase Z-DNA-binding protein Zα domain
3eyi - ZDNA hexamer CGCGCG + Z-DNA-binding protein Zβ domain
1j75 - ZDNA hexamer CGCGCG + DLM-1 Zα domain
3fqb - ZDNA hexamer CGCGTG + Ba
Additional Resources
For additional information, see: Nucleic Acids
References
- ↑ 1.0 1.1 1.2 Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003 Jul;4(7):566-72. PMID:12838348 doi:10.1038/nrg1115
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007 May 1;12:4424-38. PMID:17485386
- ↑ Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005 Oct 20;437(7062):1183-6. PMID:16237447 doi:10.1038/nature04088
- ↑ Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999 Jun 11;284(5421):1841-5. PMID:10364558
- ↑ Kim YG, Lowenhaupt K, Oh DB, Kim KK, Rich A. Evidence that vaccinia virulence factor E3L binds to Z-DNA in vivo: Implications for development of a therapy for poxvirus infection. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1514-8. Epub 2004 Feb 2. PMID:14757814 doi:10.1073/pnas.0308260100
- ↑ Drozdzal P, Gilski M, Kierzek R, Lomozik L, Jaskolski M. High-resolution crystal structure of Z-DNA in complex with Cr cations. J Biol Inorg Chem. 2015 Feb 17. PMID:25687556 doi:http://dx.doi.org/10.1007/s00775-015-1247-5
- ↑ http://203.129.231.23/indira/nacc/
- ↑ Rich A, Nordheim A, Wang AH. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791-846. PMID:6383204 doi:http://dx.doi.org/10.1146/annurev.bi.53.070184.004043
- ↑ Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680-6. PMID:514347
- ↑ Sinden, Richard R (1994-01-15). DNA structure and function (1st ed.). Academic Press. pp. 398. ISBN 0-12-645750-6.
Proteopedia Page Contributors and Editors (what is this?)
Adithya Sagar, Michal Harel, Joel L. Sussman, Alexander Berchansky, David Canner, Donald Voet, Jaime Prilusky, Karl Oberholser, Eran Hodis