We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Polygalacturonase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
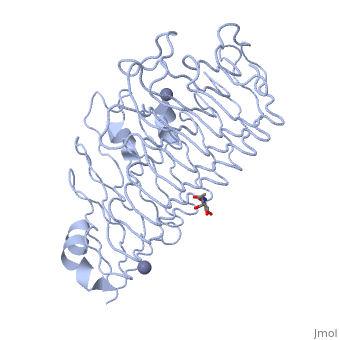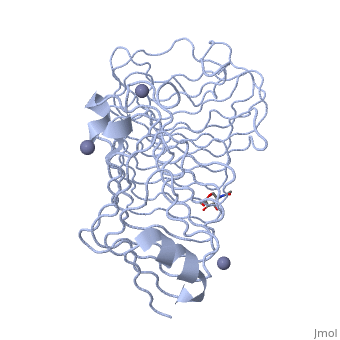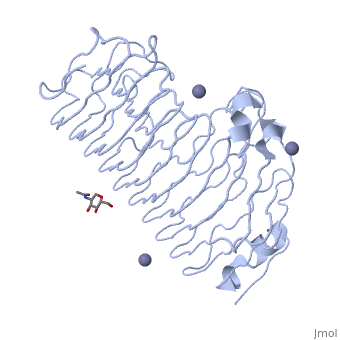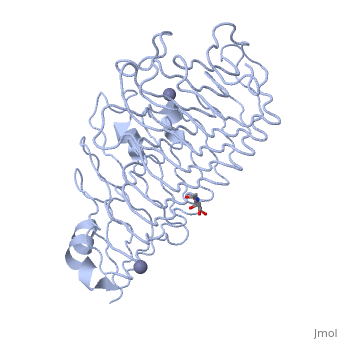
| - | <Structure load='1czf' size='350' side='right' caption='Glycosylated structure of | + | <Structure load='1czf' size='350' side='right' caption='Glycosylated structure of endo-polygalacturonase II complex with Zn+2 ion (grey) (PDB code [[1czf]])' scene=''> |
| - | + | ||
'''Polygalacturonases''' (PGs) catalyze the enzymatic depolymerization of pectates – polysaccharides that comprise the plant cell wall. Polymer disassembly of substrates by ''exo-'' and ''endo-'' PGs is carried out via a hydrolytic mechanism. Degradation of pectates in plant cell walls contributes to ripening of fruits, such as tomatoes and melons <ref>DOI: 10.1104/pp.117.2.337</ref>. Microbial PGs have been identified to be a part of defense mechanisms because of their role in pathogen attack <ref>DOI: 10.1074/jbc.273.38.24660</ref>. | '''Polygalacturonases''' (PGs) catalyze the enzymatic depolymerization of pectates – polysaccharides that comprise the plant cell wall. Polymer disassembly of substrates by ''exo-'' and ''endo-'' PGs is carried out via a hydrolytic mechanism. Degradation of pectates in plant cell walls contributes to ripening of fruits, such as tomatoes and melons <ref>DOI: 10.1104/pp.117.2.337</ref>. Microbial PGs have been identified to be a part of defense mechanisms because of their role in pathogen attack <ref>DOI: 10.1074/jbc.273.38.24660</ref>. | ||
Revision as of 19:09, 7 January 2018
| |||||||||||
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Krishna Amin, Michal Harel, Marilyn Yoder, OCA, Jaime Prilusky