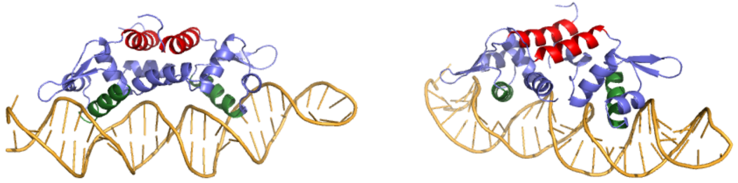We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1053
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
== Zinc Binding Site== | == Zinc Binding Site== | ||
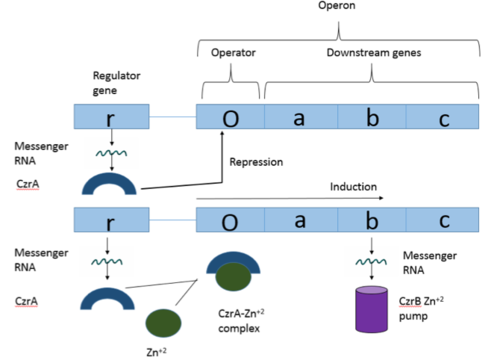
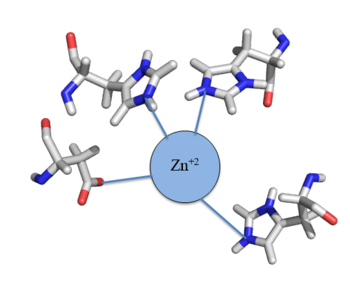
| - | Many zinc-dependent proteins are transcriptional regulators<ref>DOI: 10.1128/MMBR.00015-06</ref>. Czr A fits into this category as an [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitor] of the czr operon. Two [https://en.wikipedia.org/wiki/Zinc Zn<sup> +2</sup>] ions may bind to the dimer<ref name="critical"/>, at the location of the <scene name='69/694220/A5_helices__zn_binding/2'> | + | Many zinc-dependent proteins are transcriptional regulators<ref>DOI: 10.1128/MMBR.00015-06</ref>. Czr A fits into this category as an [https://en.wikipedia.org/wiki/Allosteric_regulation allosteric inhibitor] of the czr operon. Two [https://en.wikipedia.org/wiki/Zinc Zn<sup> +2</sup>] ions may bind to the dimer<ref name="critical"/>, at the location of the <scene name='69/694220/A5_helices__zn_binding/2'>α5 helix</scene> from each monomer. As zinc binds, the α5 helices <scene name='69/694218/2kjc_zinc_bound/1'>unalign</scene> to inhibit the DNA binding residues (Figure 2). Furthermore, CzrA must be in its dimer form for zinc to bind. The <scene name='69/694220/Spacefill_zinc_pockets/1'>zinc binding pockets</scene> are formed by two residues from each monomer, so Zn<sup>+2</sup> cannot bind to the monomer. The <scene name='69/694220/Zinc_binding_residues/7'>zinc binding site</scene> is formed by Asp84 and His 6 from one monomer, as well as His97 and His100 from the other monomer. Zinc ions were not present in the solution NMR structure<ref name="critical"/>, so a representation of a zinc ion in the binding pocket has been drawn in Figure 4. The large number of histidines used in the Czr A zinc pocket is a repetitive and commonly found feature in zinc-binding proteins <ref>Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.</ref>. |
[[Image:Zinc tetrahedral complex.PNG|350px|thumb|center| Figure 4: Zn<sup>+2</sup> tetrahedral binding complex]] | [[Image:Zinc tetrahedral complex.PNG|350px|thumb|center| Figure 4: Zn<sup>+2</sup> tetrahedral binding complex]] | ||
Revision as of 01:35, 19 January 2018
Zinc Dependent Transcriptional Repressor of the Czr operon (CzrA)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Arunkumar A., Campanello G., Giedroc D. (2009). Solution Structure of a paradigm ArsR family zinc sensor in the DNA-bound state. PNAS 106:43 18177-18182.
- ↑ Chakravorty DK, Wang B, Lee CW, Giedroc DP, Merz KM Jr. Simulations of allosteric motions in the zinc sensor CzrA. J Am Chem Soc. 2012 Feb 22;134(7):3367-76. doi: 10.1021/ja208047b. Epub 2011 Nov , 14. PMID:22007899 doi:http://dx.doi.org/10.1021/ja208047b
- ↑ MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006 Sep;70(3):583-604. PMID:16959962 doi:http://dx.doi.org/10.1128/MMBR.00015-06
- ↑ Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun 4;4(6):1609-1614.
- ↑ Grossoehme NE, Giedroc DP. Energetics of allosteric negative coupling in the zinc sensor S. aureus CzrA. J Am Chem Soc. 2009 Dec 16;131(49):17860-70. doi: 10.1021/ja906131b. PMID:19995076 doi:http://dx.doi.org/10.1021/ja906131b