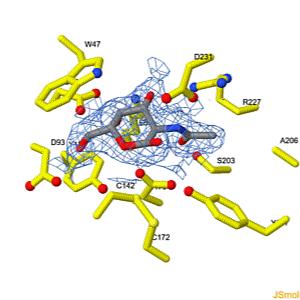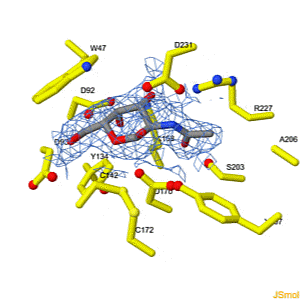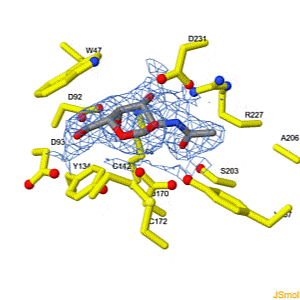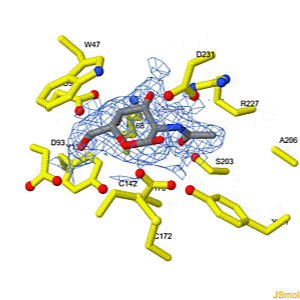
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jaime.Prilusky/Test/Sortable
From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 12: |
Line 12: |
| | </tr> | | </tr> |
| | <tr> | | <tr> |
| - | <td bgcolor="#dae4d9"> {{Proteopedia:Featured JRN/{{#expr: {{#time:U}} mod {{Proteopedia:Number of JRN articles}}}}}}</td> | + | <td style="padding: 5px;"> {{Proteopedia:Featured JRN/{{#expr: {{#time:U}} mod {{Proteopedia:Number of JRN articles}}}}}}</td> |
| - | <td bgcolor="#f1b840"> {{Proteopedia:Featured ART/{{#expr: {{#time:U}} mod {{Proteopedia:Number of ART articles}}}}}}</td> | + | <td style="padding: 5px;">{{Proteopedia:Featured ART/{{#expr: {{#time:U}} mod {{Proteopedia:Number of ART articles}}}}}}</td> |
| - | <td bgcolor="#33ff7b"> {{Proteopedia:Featured SEL/{{#expr: {{#time:U}} mod {{Proteopedia:Number of SEL articles}}}}}}</td> | + | <td style="padding: 5px;"> {{Proteopedia:Featured SEL/{{#expr: {{#time:U}} mod {{Proteopedia:Number of SEL articles}}}}}}</td> |
| - | <td bgcolor="#dae4d9"> {{Proteopedia:Featured EDU/{{#expr: {{#time:U}} mod {{Proteopedia:Number of EDU articles}}}}}}</td> | + | <td style="padding: 5px;"> {{Proteopedia:Featured EDU/{{#expr: {{#time:U}} mod {{Proteopedia:Number of EDU articles}}}}}}</td> |
| | </tr> | | </tr> |
| | </table> | | </table> |
Revision as of 05:31, 17 February 2018
|
Welcome to Proteopedia
ISSN 2310-6301
The free, collaborative 3D-encyclopedia of proteins & other molecules
|
| Journals |
Art on Science |
Selected Pages |
Education |
| |
|
|
|