Sandbox Reserved 1347
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Structural highlights == | == Structural highlights == | ||
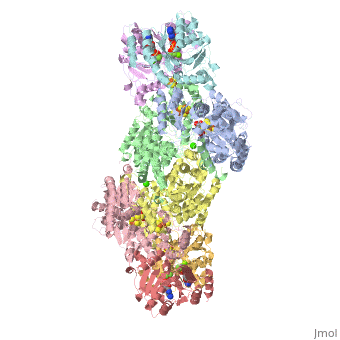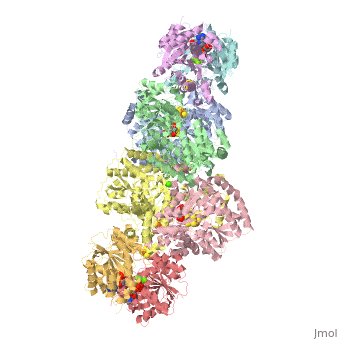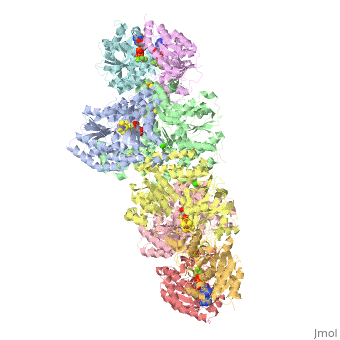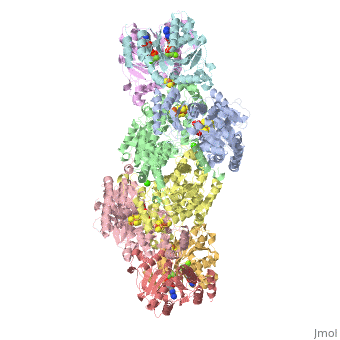
| + | The structure of nitrogenase consists of metal clusters centered throughout the protein. Three of these clusters are the 4Fe-4S Cluster, P-Cluster, and FeMo-Cluster. At either end of the protein are two copies of the Fe protein dimer, which is also the site of the ATP binding site. At these sites are ADP molecules which form a stable complex with the Fe protein. The MoFe protein, the central components of the protein, is where most of nitrogenase’ function is carried out. This protein requires a constant state of electrons which is supplied by the Fe protein which uses the hydrolysis of ATP to pump these electrons into the MoFe protein. Thus, it is helpful for the Fe protein to be coupled with ADP/ATP at the ends of the protein. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT by starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT by starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 20:32, 21 February 2018
| This Sandbox is Reserved from January through July 31, 2018 for use in the course HLSC322: Principles of Genetics and Genomics taught by Genevieve Houston-Ludlam at the University of Maryland, College Park, USA. This reservation includes Sandbox Reserved 1311 through Sandbox Reserved 1430. |
To get started:
More help: Help:Editing |
Nitrogenase
|
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644