Pho4
From Proteopedia
| Line 23: | Line 23: | ||
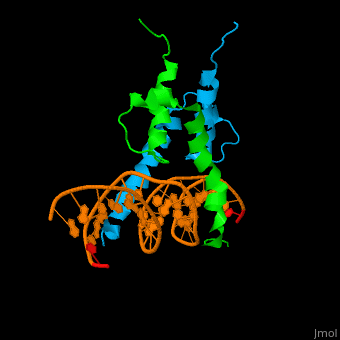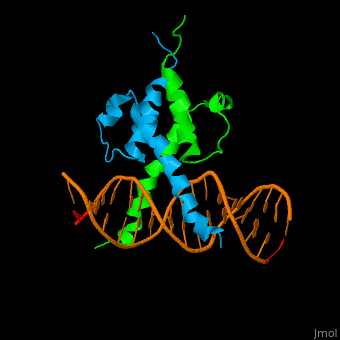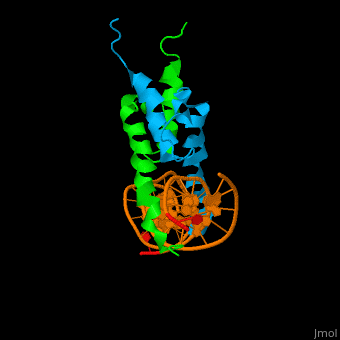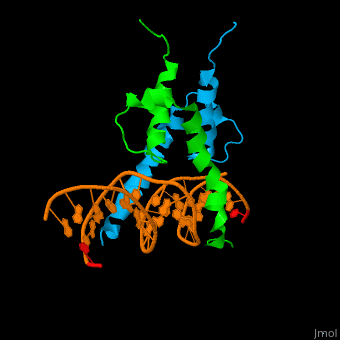
The structure of the Pho4 protein is a basic Helix-loop-helix(bHLH) , The linker sequence between Helix1(H1) and Helix2(H2) contains several residues: Asn, Glu, Pro and Ser<ref>PMID: 3775366</ref> . The DNA binding site is a homodimer and two monomers in a left‐handed four‐helix bundle fold (identical to bHLH/Zip and bHLH proteins fold) . | The structure of the Pho4 protein is a basic Helix-loop-helix(bHLH) , The linker sequence between Helix1(H1) and Helix2(H2) contains several residues: Asn, Glu, Pro and Ser<ref>PMID: 3775366</ref> . The DNA binding site is a homodimer and two monomers in a left‐handed four‐helix bundle fold (identical to bHLH/Zip and bHLH proteins fold) . | ||
| - | + | *loop structure | |
Pho4 loops (conserved structure?) are long but compact and forms a short alpha-helical structure <ref>PMID:1170095</ref>. The PHO4 loop contains a Trp residue that faces the aromatic rings of Tyr52 and His55 of helix H2, and Pro28 of H2 and H1.an aromatic cluster forms a cap structure is observed.<ref>PMID: 9303313</ref> | Pho4 loops (conserved structure?) are long but compact and forms a short alpha-helical structure <ref>PMID:1170095</ref>. The PHO4 loop contains a Trp residue that faces the aromatic rings of Tyr52 and His55 of helix H2, and Pro28 of H2 and H1.an aromatic cluster forms a cap structure is observed.<ref>PMID: 9303313</ref> | ||
Revision as of 12:38, 19 March 2018
| |||||||||||
sturcture
The structure of the Pho4 protein is a basic Helix-loop-helix(bHLH) , The linker sequence between Helix1(H1) and Helix2(H2) contains several residues: Asn, Glu, Pro and Ser[6] . The DNA binding site is a homodimer and two monomers in a left‐handed four‐helix bundle fold (identical to bHLH/Zip and bHLH proteins fold) .
- loop structure
Pho4 loops (conserved structure?) are long but compact and forms a short alpha-helical structure [7]. The PHO4 loop contains a Trp residue that faces the aromatic rings of Tyr52 and His55 of helix H2, and Pro28 of H2 and H1.an aromatic cluster forms a cap structure is observed.[8]
3D Structures of lactoferrin
Updated on 19-March-2018
1a0a – yPho4 DNA-binding domain + DNA - yeast
3w3x – yPho4 peptide + importin subunit b-3
References
- ↑ Berben G, Legrain M, Gilliquet V, Hilger F. The yeast regulatory gene PHO4 encodes a helix-loop-helix motif. Yeast. 1990 Sep-Oct;6(5):451-4. PMID:2220078 doi:http://dx.doi.org/10.1002/yea.320060510
- ↑ Berben G, Legrain M, Gilliquet V, Hilger F. The yeast regulatory gene PHO4 encodes a helix-loop-helix motif. Yeast. 1990 Sep-Oct;6(5):451-4. PMID:2220078 doi:http://dx.doi.org/10.1002/yea.320060510
- ↑ Yoshida K, Ogawa N, Oshima Y. Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1989 May;217(1):40-6. PMID:2671650
- ↑ Yoshida K, Ogawa N, Oshima Y. Function of the PHO regulatory genes for repressible acid phosphatase synthesis in Saccharomyces cerevisiae. Mol Gen Genet. 1989 May;217(1):40-6. PMID:2671650
- ↑ Tamai Y, Toh-e A, Oshima Y. Regulation of inorganic phosphate transport systems in Saccharomyces cerevisiae. J Bacteriol. 1985 Nov;164(2):964-8. PMID:3902805
- ↑ Leszczynski JF, Rose GD. Loops in globular proteins: a novel category of secondary structure. Science. 1986 Nov 14;234(4778):849-55. PMID:3775366
- ↑ Schmeckpeper BJ, Adams JM, Harris AW. Detection of a possible precursor of immunoglobulin light chain in MOPC 41 A plasmacytoma cells. FEBS Lett. 1975 Apr 15;53(1):95-8. PMID:1170095
- ↑ Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313 doi:10.1093/emboj/16.15.4689
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Inbar Yaffe, Alexander Berchansky, Joel L. Sussman