We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
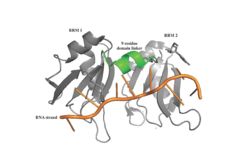
[[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | [[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | ||
| - | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the 3'poly (A) tail of mRNA that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and initiation factors causes it to also play a significant role in translation initiation and mRNA stabilization and degradation. PABP consists of four conserved domains of RNA recognition motifs (RRMs); however, the two N-terminal RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of PABP, so RRM3 and RRM4 may not be essential. Thus, the published X-ray structure exhibits RRM1 and RRM2 at a 2.6 Angstrom resolution. This is shown as <scene name='78/781947/Biological_assembly_1/1'>Biological Assembly 1</scene>. Both RRM 1 and 2 are needed to support biochemical function, that is, no one RRM can support biochemical function. Additionally, there is a proline rich C-terminal portion of variable length that is not well conserved and unknown as to how it contributes to the protein's function. <ref> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2.</ref> | + | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the 3'poly (A) tail of mRNA that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and initiation factors causes it to also play a significant role in translation initiation and mRNA stabilization and degradation. PABP consists of four conserved domains of RNA recognition motifs (RRMs); however, the two N-terminal RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of PABP, so RRM3 and RRM4 may not be essential. Thus, the published X-ray structure exhibits RRM1 and RRM2 at a 2.6 Angstrom resolution. This is shown as <scene name='78/781947/Biological_assembly_1/1'>Biological Assembly 1</scene>. Both RRM 1 and 2 are needed to support biochemical function, that is, no one RRM can support biochemical function. Additionally, there is a proline rich C-terminal portion of variable length that is not well conserved and unknown as to how it contributes to the protein's function. <ref name= "PABP"> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2.</ref> |
== Function == | == Function == | ||
| Line 23: | Line 23: | ||
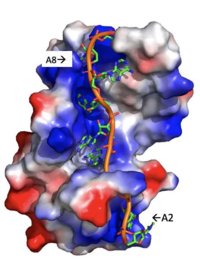
====Adenosine Stabilization Interaction Patterns==== | ====Adenosine Stabilization Interaction Patterns==== | ||
| - | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88. <ref | + | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781949/Lys_104_asp_105/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between aromatic and alipathic side chains. <scene name='78/781947/Interactions_with_a3/1'>Adenosine-3 sandwiching</scene> occurs between aromatic and alipathic side chains and is specified by Lysine 104, and <scene name='78/781947/Residues_interacting_with_a6/1'>Adenosine-6 sandwiching</scene> occurs similarly, but it is specified doubly by two residues, Trp-86 and Gln-88. <ref name= "PABP"/> |
===Translation Initiation=== | ===Translation Initiation=== | ||
| Line 29: | Line 29: | ||
The initiation of translation in eukaryotes requires many translation factors and proteins, one of which is PABP. There is evidence that PABP is critical for formation of the “closed loop” model of protein synthesis, which involves joining the 3’ poly (A) tail of mRNA to the 5’ cap to create circular RNA. (B) This process utilizes eIF4F, a protein composed of multiple TFs that play various roles in translation. eIF4G is a scaffolding protein that binds the other subunits, eIF4E and eIF4A. eIF4E creates interactions with the 5’ cap to bring the IF complex to the 5’ end of the mRNA. eIF4A is an RNA helicase that denatures RNA and allows the ribosome to move along the strand. (C) All of these proteins are known to be involved in protein synthesis, but several mechanisms have been proposed for how eIF4G might also be interacting with PABP. | The initiation of translation in eukaryotes requires many translation factors and proteins, one of which is PABP. There is evidence that PABP is critical for formation of the “closed loop” model of protein synthesis, which involves joining the 3’ poly (A) tail of mRNA to the 5’ cap to create circular RNA. (B) This process utilizes eIF4F, a protein composed of multiple TFs that play various roles in translation. eIF4G is a scaffolding protein that binds the other subunits, eIF4E and eIF4A. eIF4E creates interactions with the 5’ cap to bring the IF complex to the 5’ end of the mRNA. eIF4A is an RNA helicase that denatures RNA and allows the ribosome to move along the strand. (C) All of these proteins are known to be involved in protein synthesis, but several mechanisms have been proposed for how eIF4G might also be interacting with PABP. | ||
| - | By observing protein synthesis in cells deficient of PABP, <ref name="kahvejian">Kahvejian, A. “Mammalian Poly(A)-Binding Protein Is a Eukaryotic Translation Initiation Factor, Which Acts via Multiple Mechanisms.” Genes & Development, vol. 19, no. 1, 2005, pp. 104–113., doi:10.1101/gad.1262905</ref> | + | By observing protein synthesis in cells deficient of PABP, Kahvejian et. al <ref name="kahvejian">Kahvejian, A. “Mammalian Poly(A)-Binding Protein Is a Eukaryotic Translation Initiation Factor, Which Acts via Multiple Mechanisms.” Genes & Development, vol. 19, no. 1, 2005, pp. 104–113., doi:10.1101/gad.1262905</ref> were able to show that the PABP/eIF4G interaction promotes translation. The cells lacking PABP showed a seven-fold decrease in the rate of translation, which was remedied by reintroducing PABP to the cells. Other cells were treated with a PABP mutant that also had an eIF4G binding site, but the introduction of these proteins did not return the rate of translation to its normal level (C) These results show that not only is PABP acting as a TF in eukaryotic cells, but it also needs to interact with eIF4G in order to have an effect. |
<scene name='78/781946/Pabp_linker_conserved_residues/1'>PABP linker with Conserved Residues Shown</scene> | <scene name='78/781946/Pabp_linker_conserved_residues/1'>PABP linker with Conserved Residues Shown</scene> | ||
Revision as of 18:30, 29 March 2018
Poly(A) binding protein
Structure
| |||||||||||