We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 18: | Line 18: | ||
[[Image: Stacking_of_adenosines_with_beta_sheets.jpg |250 px|left|thumb|Figure 2: The specific weak intermolecular interactions between RNP1 and RNP2 and Adenosines. These interactions are the primary support of adenosine recognition by PABP and include mainly van der Waals interactions, hydrogen bonds, and stacking interactions. ]] | [[Image: Stacking_of_adenosines_with_beta_sheets.jpg |250 px|left|thumb|Figure 2: The specific weak intermolecular interactions between RNP1 and RNP2 and Adenosines. These interactions are the primary support of adenosine recognition by PABP and include mainly van der Waals interactions, hydrogen bonds, and stacking interactions. ]] | ||
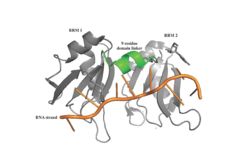
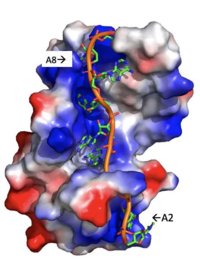
| - | A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by [https://en.wikipedia.org/wiki/Electrophoretic_mobility_shift_assay EMSA competition experiments], there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for [https://en.wikipedia.org/wiki/Crystallization crystallization] and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel [https://en.wikipedia.org/wiki/Beta_sheet β-strands] and two [https://en.wikipedia.org/wiki/Alpha_helix α-helices]. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two [https://en.wikipedia.org/wiki/Conserved_sequence conserved sequences] in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 [https://en.wikipedia.org/wiki/Residue residues], while RNP2 consists of a conserved sequence of 6 residues. Much of the weak [https://en.wikipedia.org/wiki/Intermolecular_force intermolecular interactions] with [https://en.wikipedia.org/wiki/Adenosine adenosine] from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. <ref name= "PABP"/> | + | A primary function of PABP is recognizing and interacting with the 3'poly (A) tail created in mRNA processing. As found by [https://en.wikipedia.org/wiki/Electrophoretic_mobility_shift_assay EMSA competition experiments], there is a minimum of 11-12 adenosines necessary in the poly (A) tail for the adenosine chain to bind to PABP with high affinity. However, for one biological assembly, a chain containing 9 adenosines sufficiently binds the assembly for [https://en.wikipedia.org/wiki/Crystallization crystallization] and is shown in the biological assembly structure. The 4 RRM domains that are the primary interacting sites for the adenosine recognition exist as globular domains, each having four antiparallel [https://en.wikipedia.org/wiki/Beta_sheet β-strands] and two [https://en.wikipedia.org/wiki/Alpha_helix α-helices]. With the N-terminal to C-terminal motifs labeled as S1 to S4 for the β-strands and H1 to H2 for the α-helices, the strands are spatially arranged as S2-S3-S1-S4. Furthermore, there are two [https://en.wikipedia.org/wiki/Conserved_sequence conserved sequences] in each RRM, called RNP1 and 2. RNP 1 consists of a conserved sequence of 8 [https://en.wikipedia.org/wiki/Residue residues], while RNP2 consists of a conserved sequence of 6 residues. Much of the weak [https://en.wikipedia.org/wiki/Intermolecular_force intermolecular interactions] with [https://en.wikipedia.org/wiki/Adenosine adenosine] from the RRMs occur from the <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2</scene> conserved sequences, which correspond to the two central β-strands, with specific interactions shown in Figure 2.The support for adenosine recognition by the RRMs occurs as a type of binding trough with the sheets, primarily <scene name='78/781946/Rnp1_rnp2_primary_a_binding/1'>RNP1 and RNP2 forming the Primary Binding Trough</scene>, and the interstrand loop between β-strands 2 and 3 as well as the domain linker forming the <scene name='78/781946/Adenosine_binding_wall/1'> Adenosine Binding Wall</scene>. It is believed that the <scene name='78/781946/Pabp_linker_conserved_residues/1'>Conserved Linker Shown</scene> is disordered in the absence of RNA because it does not intramolecularly interact with either of the RRM motifs. Additionally, the primary binding trough is stabilized by <scene name='78/781946/Rrm1_2_packing_intxn/2'>Stabilizing Packing Interactions of RRM1 RRM2 Binding Trough</scene>. <ref name= "PABP"/> |
[[Image: Adenosine_backbone.png |200 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 (shown in blue) make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | [[Image: Adenosine_backbone.png |200 px|right|thumb|Figure 3: Basic residues of RRM 1 and 2 (shown in blue) make stabilizing electrostatic interactions with the negatively charged adenosine phosphates. ]] | ||
| Line 27: | Line 27: | ||
===Translation Initiation=== | ===Translation Initiation=== | ||
| - | The initiation of translation in eukaryotes requires many translation factors and proteins, one of which is PABP. There is evidence that PABP is critical for formation of the “closed loop” model of protein synthesis, which involves joining the 3’ poly (A) tail of mRNA to the 5’ cap to create circular RNA. ( | + | The initiation of translation in eukaryotes requires many translation factors and proteins, one of which is PABP. There is evidence that PABP is critical for formation of the “closed loop” model of protein synthesis, which involves joining the 3’ poly (A) tail of mRNA to the 5’ cap to create circular RNA <ref name="imataka"> Imataka, H. “A Newly Identified N-Terminal Amino Acid Sequence of Human eIF4G Binds Poly(A)-Binding Protein and Functions in Poly(A)-Dependent Translation.” The EMBO Journal, vol. 17, no. 24, 1998, pp. 7480–7489., doi:10.1093/emboj/17.24.7480.</ref>. This process utilizes eIF4F, a protein composed of multiple TFs that play various roles in translation. eIF4G is a scaffolding protein that binds the other subunits, eIF4E and eIF4A. eIF4E creates interactions with the 5’ cap to bring the IF complex to the 5’ end of the mRNA. eIF4A is an RNA helicase that denatures RNA and allows the ribosome to move along the strand. eIF4G also has a binding site for PABP, which is N-terminal to the binding site for eIF4F and interacts with the same RRMs that allow PABP to bind RNA. <ref name="Osvaldo">De Melo Neto, Osvaldo P., et al. “Phosphorylation and Interactions Associated with the Control of the Leishmania Poly-A Binding Protein 1 (PABP1) Function during Translation Initiation.” RNA Biology, 23 Mar. 2018, pp. 1–17., doi:10.1080/15476286.2018.1445958.</ref> All of these proteins are known to be involved in protein synthesis, but several mechanisms have been proposed for how PABP might be promoting translation. |
| - | By observing protein synthesis in cells deficient of PABP, Kahvejian et al. were able to show that the PABP/eIF4G interaction promotes translation. The cells lacking PABP showed a seven-fold decrease in the rate of translation, which was remedied by reintroducing PABP to the cells. Other cells were treated with a PABP mutant that also had an eIF4G binding site, but the introduction of these proteins did not return the rate of translation to its normal level ( | + | By observing protein synthesis in cells deficient of PABP, Kahvejian et al. were able to show that the PABP/eIF4G interaction promotes translation. The cells lacking PABP showed a seven-fold decrease in the rate of translation, which was remedied by reintroducing PABP to the cells. Other cells were treated with a PABP mutant that also had an eIF4G binding site, but the introduction of these proteins did not return the rate of translation to its normal level <ref name="kahvejian"> Kahvejian, A. “Mammalian Poly(A)-Binding Protein Is a Eukaryotic Translation Initiation Factor, Which Acts via Multiple Mechanisms.” Genes & Development, vol. 19, no. 1, 2005, pp. 104–113., doi:10.1101/gad.1262905.</ref>. These results show that not only is PABP acting as a TF in eukaryotic cells, but it also needs to interact with eIF4G in order to have an effect. |
| - | Furthermore, PABP interactions have been shown to be critical for the binding of the 80S ribosomal subunit. In a similar experiment, cells that were deficient in PABP were observed for binding of the 80S subunit, and researchers saw a binding reduction of greater than 60%. Reintroducing PABP to these cells restored ribosomal binding and even promoted the formation of the 80S initiation complex. These results display the importance of PABP in recruiting the 80S ribosomal subunit for the initiation of translation. A similar experiment was run In order to determine whether decreased 80S ribosomal recruitment was due to a decrease in 40S ribosomal recruitment. Results showed that PABP is also involved in recruiting the 40S ribosomal subunit to the RNA | + | Furthermore, PABP interactions have been shown to be critical for the binding of the 80S ribosomal subunit. In a similar experiment, cells that were deficient in PABP were observed for binding of the 80S subunit, and researchers saw a binding reduction of greater than 60%. Reintroducing PABP to these cells restored ribosomal binding and even promoted the formation of the 80S initiation complex. These results display the importance of PABP in recruiting the 80S ribosomal subunit for the initiation of translation. A similar experiment was run In order to determine whether decreased 80S ribosomal recruitment was due to a decrease in 40S ribosomal recruitment. Results showed that PABP is also involved in recruiting the 40S ribosomal subunit to the RNA <ref name="kahvejian">. Because of this, PABP affects the binding of the 80S subunit in two ways: indirectly, by allowing the 40S subunit to be available to bind, and directly, by promoting association of the 80S subunit with the 40S subunit. |
| + | In addition to these interactions, the PABP aids in this closed loop formation via interactions with Poly (A) Interacting Protein 1 (PAIP-1). In this process, PABP interacts with PAIP-1 to facilitate the complex to interact with eIF4A initiation factor. eIF4A unwinds the 5' untranslated region (UTR) of an mRNA transcript, which signals eIF4G to function as previously mentioned <ref name="PABP"/>. Thus, there are two mechanisms to initiate this loop closing pathway with eIF4G. | ||
| - | <scene name='78/ | + | ====Structural Components of PABP Translation Initiation==== |
| + | |||
| + | The RRMs support interactions with the interacting proteins such as eIF4G and PAIP-1, however, the specific ways in which PABP interacts with these proteins are not structurally proven. However, there is a convex dorsal surface present on the RRM 1 and 2 motifs formed by the two α-helices in each RRM, specified as H1 and H2 in RRM1 and H1' and H2' in RRM2. This surface contains a sequence portion of <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Conserved Hydrophobic Residues</scene>and <scene name='78/781947/Hydrophilic_residues/2'>Conserved Acidic Residues</scene> | ||
| + | residues. It is thought that this area of conservation interacts with such translation factors and regulating proteins. For example, the conserved acidic residues may be beneficial to be used in to interact with essential basic residues present in both eIF4G and PAIP-1 via ionic interactions <ref name="PABP"/>. | ||
| - | <scene name='78/781947/H1_and_h2_h2ophobic_residues/2'>Phe 74 Phe 122 Met 158 Met 161 Leu163</scene> | ||
| - | <scene name='78/781947/Hydrophilic_residues/2'>Asp70 Asp111 Asp117</scene> | ||
<scene name='78/781947/Pro-ser_in_linker/2'>Pro91Ser92</scene> | <scene name='78/781947/Pro-ser_in_linker/2'>Pro91Ser92</scene> | ||
Revision as of 19:25, 29 March 2018
Poly(A) binding protein
Structure
| |||||||||||