User:Ben Dawson/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Background == | == Background == | ||
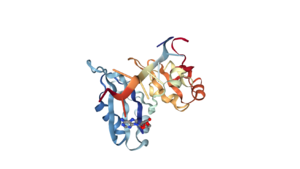
<Structure load='1CVJ' size='350' frame='true' align='right' caption='Poly-A Binding Protein (PABP)' scene='Insert optional scene name here' /> | <Structure load='1CVJ' size='350' frame='true' align='right' caption='Poly-A Binding Protein (PABP)' scene='Insert optional scene name here' /> | ||
| + | The Human Poly-A Binding Protein (PABP) is a mRNA binding protein that binds to the 3’ Poly-A tail on mRNA. Through extensive Adenosine recognition by the RNA REcognition Motifs (RRMs) of PABP, the protein is involved in three main functions: recognition of the 3’ Poly-A tail, mRNA stabilization, and eukaryotic translation initiation. The contributions of controlling gene expression via different families of PABPs is not yet fully understood. PABP families are divided into nuclear and cytoplasmic.5 PABP1, which is predominantly cytoplasmic, is often referred to as PABP because it is the only form of PABP that has been extensively studied in its role with mRNA translation and stability.5 | ||
| + | |||
| + | |||
== Structure == | == Structure == | ||
| + | |||
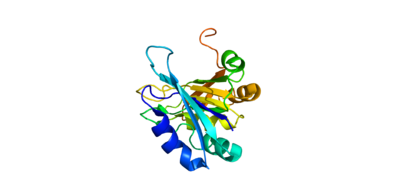
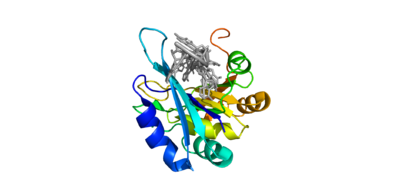
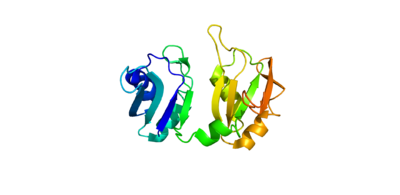
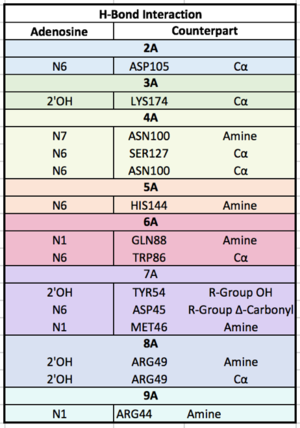
| + | The crystall structure was derived from X-ray Diffraction at 2.6Å (R-value: 23%). It is comprised of four RNA recognition motifs (RRMs), which are highly conserved RNA-binding domains.¹ The RRM in PABP is found in over two hundred families of proteins across species, indicating that it is ancient.¹ RRM1 and 2 are examined in this article. | ||
| + | Each RRM has four-stranded antiparallel beta sheet and two helices.¹ Recent studies have shown that mRNA binding is due to the presence of the beta-sheet surfaces.² (image- trough). The beta-sheet flooring present in PABP interacts with the 3’ mRNA tail via a combination of van der Waals, aromatic stacking, and Hydrogen bonding. Through these interactions, PABP binds to 3’ poly-adenosine tail with a KD of 2-7 nM.5 The structural elements highlighted consist of the RRM1/2 subunits, the linker domain, and the Poly-A mRNA binding trough. | ||
| + | |||
===RNA Recognition Motifs (RRMs)=== | ===RNA Recognition Motifs (RRMs)=== | ||
| Line 26: | Line 33: | ||
== Function == | == Function == | ||
| + | Polyadenylation of an mRNA involves the recognition of the 5’-AAUAAA-3’ consensus site, the cleavage downstream of the consensus site, and then the addition of adenines by Poly-A Polymerase to the 3’ end. The newly added Poly-A tail is associated with the Poly-A Binding Protein (PABP), working together to stabilize mRNA by preventing exoribonucleolytic degradation,¹ thereby guiding the mRNA molecule into the translation pathway. Upon mRNA poly-A recognition, PABP and the bound mRNA stimulate the initiation of translation by interacting with initiation factor eIF4G. | ||
| + | |||
| + | PABP and mRNA complex aids in translation initiation under two proposed mechanisms. Within the two mechanisms, studies have highlighted the presence The “Closed Loop” Model entails the recognition of the 5’ 7-methyl-Guanosine Cap by eIF4F, which is a ternary complex made up of a cap-binding protein (eIF4E) and RNA helicase (eIF4A) connected by the bridging protein (eIF4G).¹ Translation initiation is stimulated by the PABP bound to the Poly-A tail and its association with eIF4G.¹ The 5’ UTR is unwound by the elF4F complex, and ribosomes are recruited to create the initiation complex. The eIF4G protein then guides the 40S subunit to the start codon (AUG), which is followed by the binding 60S ribosomal subunit, creating the 80S initiation complex.¹ The association of the PolyA binding protein and eIF4G gave rise to the name “closed loop.”¹ | ||
| + | |||
| + | In more complex eukaryotic organisms, PABP indirectly stimulates translation via PAIP-1 (PABP interacting protein). A higher presence of PAIP-1 increases the rate of translation initiation, indicating another way to “close the loop.”¹ | ||
| + | |||
| + | |||
== Disease == | == Disease == | ||
Revision as of 18:26, 30 March 2018
Human Poly A-Binding Protein (1CVJ)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644