User:Neel Bhagat/Sandbox 1
From Proteopedia
| Line 3: | Line 3: | ||
=== Overview === | === Overview === | ||
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM). | The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM). | ||
| + | |||
=== History === | === History === | ||
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins ([https://en.wikipedia.org/wiki/Heterogeneous_ribonucleoprotein_particle hnRNPs]). | Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins ([https://en.wikipedia.org/wiki/Heterogeneous_ribonucleoprotein_particle hnRNPs]). | ||
| Line 9: | Line 10: | ||
The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006). | The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006). | ||
<StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | <StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | ||
| + | |||
=== Structure Determination === | === Structure Determination === | ||
| Line 14: | Line 16: | ||
== Splicing Activity == | == Splicing Activity == | ||
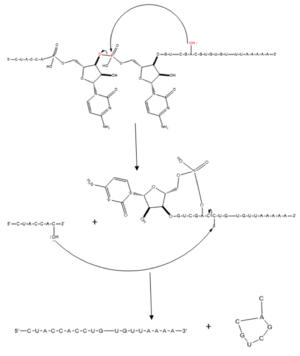
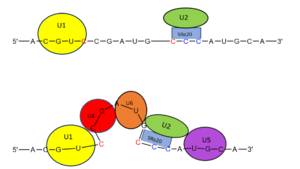
| - | The splicing mechanism for SRp20 follows the normal eukaryotic mechanism, in which five different [https://en.wikipedia.org/wiki/SnRNP small nuclear ribonucleoproteins] (snRNPs) bring the splice sites together in order to start the reaction (Figure 1). Specifically, SRp20 and other SR proteins interact with the RNA ligand at the [https://en.wikipedia.org/wiki/Exonic_splicing_enhancer exonic splicing enhancer sequence] at the beginning of the 3’ splice site adjacent to the intron being removed. SRp20 facilitates the interaction of the U2 snRNP with the RNA to continue the mechanism (Figure 2). [[Image:mechanism1.png|300px|left|thumb|'''Figure 1.''' Splicing mechanism for eukaryotes. Free 3’OH nucleophile of adenosine in intron attacks phosphorus of phosphate creating a ring structure intron known as a lariat. The free 2’OH in the 5’ splice site (red) acts as the nucleophile to attack the phosphate of the first nucleotide in the 3’ splice site (red) to release the lariat intron. The products include the final modified RNA sequence and the lariat which will be recycled.jpg]] [[Image:prettymechanism.png|300px|right|thumb|'''Figure 2.''' SRp20 works with U2 snRNP: Five snRNPs are needed in the eukaryotic splicing mechanism to facilitate the reaction. The U2 snRNP must attach to the 3’ splice site to bring together the 5’ and 3’ splice sites (red). SRp20 facilitates binding of U2 to the 3’ splice site by binding to the exonic splicing enhancer sequence (blue) in the RNA at the backbone. U2 must bind before the other snRNPs can bind to continue the mechanism.jpg]] | + | The splicing mechanism for SRp20 follows the normal eukaryotic mechanism, in which five different [https://en.wikipedia.org/wiki/SnRNP small nuclear ribonucleoproteins] (snRNPs) bring the splice sites together in order to start the reaction (Figure 1). Specifically, SRp20 and other SR proteins interact with the RNA ligand at the [https://en.wikipedia.org/wiki/Exonic_splicing_enhancer exonic splicing enhancer sequence] at the beginning of the 3’ splice site adjacent to the intron being removed. SRp20 facilitates the interaction of the U2 snRNP with the RNA to continue the mechanism (Figure 2). [[Image:mechanism1.png|300px|left|thumb|'''Figure 1.''' Splicing mechanism for eukaryotes. Free 3’OH nucleophile of adenosine in intron attacks phosphorus of phosphate creating a ring structure intron known as a lariat. The free 2’OH in the 5’ splice site (red) acts as the nucleophile to attack the phosphate of the first nucleotide in the 3’ splice site (red) to release the [https://news.brown.edu/articles/2012/06/lariats lariat] intron. The products include the final modified RNA sequence and the lariat which will be recycled.jpg]] [[Image:prettymechanism.png|300px|right|thumb|'''Figure 2.''' SRp20 works with U2 snRNP: Five snRNPs are needed in the eukaryotic splicing mechanism to facilitate the reaction. The U2 snRNP must attach to the 3’ splice site to bring together the 5’ and 3’ splice sites (red). SRp20 facilitates binding of U2 to the 3’ splice site by binding to the exonic splicing enhancer sequence (blue) in the RNA at the backbone. U2 must bind before the other snRNPs can bind to continue the mechanism.jpg]] |
| Line 35: | Line 37: | ||
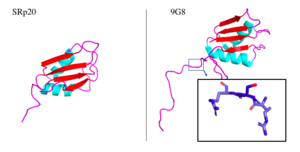
9G8 is another SR protein that is 80% similar in amino acid sequence. They are two of the smallest proteins in the SR family and both contain an RRM that promotes export of mRNA through interaction with the TAP protein. The only other protein shown to promote transport through the TAP protein is SF2, one of the first SR proteins discovered (Huang 2003). Both RRMs adopt a βαββαβ mentioned earlier. Both RRMs interact with RNA with limited selectivity and therefore recognize many different RNA sequences. The 9G8 RRM contains an large hydrophobic core on its B-sheet. Observing the 9G8 protein has proved useful in understanding SRp20 less stable protein structure. In fact, the one of the only significant structural difference between the two proteins lies in the loops between a-helix 2 and B-4 where several amino acids are not conserved. | 9G8 is another SR protein that is 80% similar in amino acid sequence. They are two of the smallest proteins in the SR family and both contain an RRM that promotes export of mRNA through interaction with the TAP protein. The only other protein shown to promote transport through the TAP protein is SF2, one of the first SR proteins discovered (Huang 2003). Both RRMs adopt a βαββαβ mentioned earlier. Both RRMs interact with RNA with limited selectivity and therefore recognize many different RNA sequences. The 9G8 RRM contains an large hydrophobic core on its B-sheet. Observing the 9G8 protein has proved useful in understanding SRp20 less stable protein structure. In fact, the one of the only significant structural difference between the two proteins lies in the loops between a-helix 2 and B-4 where several amino acids are not conserved. | ||
Aside from the RRM, both proteins have one SR-rich domain although 9G8 includes about 40 more amino acids this domain (Corbo 2013). [[Image:9G8srp20comparison.png|300px|right|thumb|'''Figure 5.''' SRp20 and 9G8 proteins. The PDB file for SRp20 (left; PDB file: 2i2y) does not include the SR-rich domain but 9G8 (right; PDB file: 2hvz) does. Boxed image shows an RSR region in the PG8 protein with oxygens highlighted in red. Images taken from PyMol software.jpg]] Within the bigger 9G8 protein, there is a zinc knuckle that allows for binding of pyrimidine-rich RNA sequences. This zinc knuckle is not present in SRp20, lending the protein to binding of more purine-rich sequences (Huang 2003).Not only are these two proteins similar, but they also play similar roles in mRNA export out of the nucleus. Both move continuously between the nucleus and cytoplasm which requires phosphorylation of its serine residues located in the SR-rich domain. Serine phosphorylation has been shown to have great importance in the proper functioning of many SR proteins (Figure Shepard & Hertel 2009). | Aside from the RRM, both proteins have one SR-rich domain although 9G8 includes about 40 more amino acids this domain (Corbo 2013). [[Image:9G8srp20comparison.png|300px|right|thumb|'''Figure 5.''' SRp20 and 9G8 proteins. The PDB file for SRp20 (left; PDB file: 2i2y) does not include the SR-rich domain but 9G8 (right; PDB file: 2hvz) does. Boxed image shows an RSR region in the PG8 protein with oxygens highlighted in red. Images taken from PyMol software.jpg]] Within the bigger 9G8 protein, there is a zinc knuckle that allows for binding of pyrimidine-rich RNA sequences. This zinc knuckle is not present in SRp20, lending the protein to binding of more purine-rich sequences (Huang 2003).Not only are these two proteins similar, but they also play similar roles in mRNA export out of the nucleus. Both move continuously between the nucleus and cytoplasm which requires phosphorylation of its serine residues located in the SR-rich domain. Serine phosphorylation has been shown to have great importance in the proper functioning of many SR proteins (Figure Shepard & Hertel 2009). | ||
| + | |||
== Medical Significance == | == Medical Significance == | ||
| + | |||
=== Cancer === | === Cancer === | ||
SRp20 has been linked to cancer in many instances, as well as other AS proteins. SRp20 has been seen to activate the AS of the CD44 adhesion molecule. SRp20 facilitates the splicing of exon v9, one important for function of CD44. Loss of function of SRp20 leading to loss of function of CD44 will lead to loss of “stickiness” of cells, a way that cancer cells can spread to other areas of the body. SRp20 can also affect the alternative splicing of oncogenes and tumor suppressors. Expression of the signaling pathway for SRp20 translation is directly triggered by oncogenic signalling pathways. For example, the Wnt pathway that is associated with cancer development also drives further expression of SRp20 and other SR proteins (7). These proteins aid in cell growth and proliferation, in that they increase expression of proteins they are involved in splicing. One study found that silencing SRp20 slowed cell proliferation, further supporting the idea that SRp20 helps with this process (8). Cell cycle regulator proteins FoxM1, Cdc25B, and PLK1 are alternatively spliced by SRp20, specifically regulating the G2/M cycles. These proteins are also associated with regulation of cell apoptosis. When these genes are overexpressed due to overexpressed SRp20, cells will no longer be able to apoptose when they are damaged, and the cell cycle will continue without regulation, causing overactive cell proliferation and eventually cancer. SRp20 has been seen to be overexpressed in ovarian, lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney cancer tissue (9). SRp20 also regulates genes associated with cellular senescence, or cellular immortality, which contributes to cancer formation. Specifically, SRp20 alternatively splices the TP53 gene, which generates the p53 senescence protein. | SRp20 has been linked to cancer in many instances, as well as other AS proteins. SRp20 has been seen to activate the AS of the CD44 adhesion molecule. SRp20 facilitates the splicing of exon v9, one important for function of CD44. Loss of function of SRp20 leading to loss of function of CD44 will lead to loss of “stickiness” of cells, a way that cancer cells can spread to other areas of the body. SRp20 can also affect the alternative splicing of oncogenes and tumor suppressors. Expression of the signaling pathway for SRp20 translation is directly triggered by oncogenic signalling pathways. For example, the Wnt pathway that is associated with cancer development also drives further expression of SRp20 and other SR proteins (7). These proteins aid in cell growth and proliferation, in that they increase expression of proteins they are involved in splicing. One study found that silencing SRp20 slowed cell proliferation, further supporting the idea that SRp20 helps with this process (8). Cell cycle regulator proteins FoxM1, Cdc25B, and PLK1 are alternatively spliced by SRp20, specifically regulating the G2/M cycles. These proteins are also associated with regulation of cell apoptosis. When these genes are overexpressed due to overexpressed SRp20, cells will no longer be able to apoptose when they are damaged, and the cell cycle will continue without regulation, causing overactive cell proliferation and eventually cancer. SRp20 has been seen to be overexpressed in ovarian, lung, breast, stomach, skin, bladder, colon, liver, thyroid, and kidney cancer tissue (9). SRp20 also regulates genes associated with cellular senescence, or cellular immortality, which contributes to cancer formation. Specifically, SRp20 alternatively splices the TP53 gene, which generates the p53 senescence protein. | ||
Revision as of 17:36, 2 April 2018
Contents |
Introduction
Overview
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other eukaryotes. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM).
History
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. Alternative splicing allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins (hnRNPs). The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the SF2 (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during B-cell development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006).
| |||||||||||