This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Ben Dawson/Sandbox1
From Proteopedia
| Line 5: | Line 5: | ||
== Background == | == Background == | ||
| - | The Human Poly(A) Binding Protein (PABP) was discovered in 1973 by the use of a sedimentation profile detailing the RNase digestion differentiated the PABP protein.6 Attempts to purify the 75 kDa protein then followed. In 1983, then considered “poly(A)-organizing protein,” was determined and purified by molecular weight, ligand-binding affinity, and amounts found in cytoplasmic portions of cell with ability to bind to free poly(A).8 The protein’s structure was initially determined at 2.6 Å as mRNA and RRM 1 & 2 yielded high cocrystal quality with 1:1 protein and ligand.1 The protein has a homopolymeric structure, containing four RNA recognition motifs (RRMs), which are conserved.7 RRM1 and RRM2 are N-terminal domains that are connected by a linker.1 Opposed to their counterparts, RRM3 and RRM4 bind Poly-A RNA less tightly than RRM1 and RRM2.1 | + | The Human Poly(A) Binding Protein ([PABP]http://https://www.rcsb.org/structure/1cvj&sa=D&ust=1522706780526000&usg=AFQjCNG3p5amSteG8OdxVyeIlGt97nCiyA) was discovered in 1973 by the use of a sedimentation profile detailing the RNase digestion differentiated the PABP protein.6 Attempts to purify the 75 kDa protein then followed. In 1983, then considered “poly(A)-organizing protein,” was determined and purified by molecular weight, ligand-binding affinity, and amounts found in cytoplasmic portions of cell with ability to bind to free poly(A).8 The protein’s structure was initially determined at 2.6 Å as mRNA and RRM 1 & 2 yielded high cocrystal quality with 1:1 protein and ligand.1 The protein has a homopolymeric structure, containing four RNA recognition motifs (RRMs), which are conserved.7 RRM1 and RRM2 are N-terminal domains that are connected by a linker.1 Opposed to their counterparts, RRM3 and RRM4 bind Poly-A RNA less tightly than RRM1 and RRM2.1 |
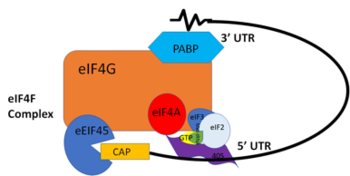
PABP is a mRNA binding protein that binds to the 3’ Poly(A) tail on mRNA. Through extensive Adenosine recognition by the RRMs of PABP, the protein is involved in three main functions: recognition of the 3’ Poly(A) tail, mRNA stabilization, and eukaryotic translation initiation. The contributions of controlling gene expression via different families of PABPs is not yet fully understood. PABP families are divided into nuclear and cytoplasmic.5 PABP1, which is predominantly cytoplasmic, is often referred to as PABP because it is the only form of PABP that has been extensively studied in its role with mRNA translation and stability.5 | PABP is a mRNA binding protein that binds to the 3’ Poly(A) tail on mRNA. Through extensive Adenosine recognition by the RRMs of PABP, the protein is involved in three main functions: recognition of the 3’ Poly(A) tail, mRNA stabilization, and eukaryotic translation initiation. The contributions of controlling gene expression via different families of PABPs is not yet fully understood. PABP families are divided into nuclear and cytoplasmic.5 PABP1, which is predominantly cytoplasmic, is often referred to as PABP because it is the only form of PABP that has been extensively studied in its role with mRNA translation and stability.5 | ||
Revision as of 21:41, 2 April 2018
Human Poly(A) Binding Protein (1CVJ)
| |||||||||||
References
1. Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell 98:6. (1999) 835-845. Print.
2. Wang, Zuoren and Kiledjian, Megerditch. “The Poly(A)-Binding Protein and an mRNA Stability Protein Jointly Regulate an Endoribonuclease Activity.” Molecular and Cellular Biology 20.17 (2000): 6334–6341. Print.
3. “Oculopharyngeal Muscular Dystrophy.” NORD (National Organization for Rare Disorders), rarediseases.org/rare-diseases/oculopharyngeal-muscular-dystrophy/.
4. Richard, Pascale, et al. “Correlation between PABPN1 Genotype and Disease Severity in Oculopharyngeal Muscular Dystrophy.” Neurology, vol. 88, no. 4, 2016, pp. 359–365., doi:10.1212/wnl.0000000000003554.
5. Gorgoni, Barbra, and Gray, Nicola. “The Roles of Cytoplasmic Poly(A)-Binding Proteins in Regulating Gene Expression: A Developmental Perspective.” Briefings in Functional Genomics and Proteomics, vol. 3, no. 2, 1 Aug. 2004, pp. 125–141., doi:10.1093/bfgp/3.2.125.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644