User:Neel Bhagat/Sandbox 1
From Proteopedia
| Line 2: | Line 2: | ||
=== Overview === | === Overview === | ||
| - | The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM). | + | The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM)(Corbo, 2013). |
=== History === | === History === | ||
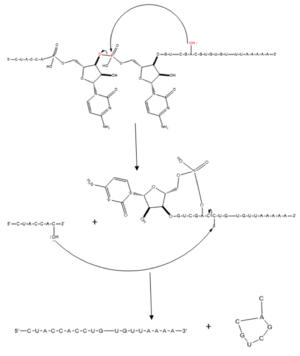
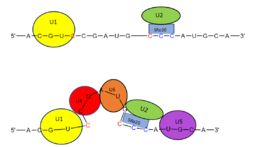
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins ([https://en.wikipedia.org/wiki/Heterogeneous_ribonucleoprotein_particle hnRNPs]). | Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. [https://en.wikipedia.org/wiki/Alternative_splicing Alternative splicing] allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins ([https://en.wikipedia.org/wiki/Heterogeneous_ribonucleoprotein_particle hnRNPs]). | ||
The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the [https://en.wikipedia.org/wiki/Serine/arginine-rich_splicing_factor_1 SF2] (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). | The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the [https://en.wikipedia.org/wiki/Serine/arginine-rich_splicing_factor_1 SF2] (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). | ||
| - | An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during [https://en.wikipedia.org/wiki/B_cell B-cell] development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). | + | An identical protein, called [http://www.uniprot.org/uniprot/Q9V3V0 X16], was discovered in an earlier paper studying different genes that change expression during [https://en.wikipedia.org/wiki/B_cell B-cell] development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). |
| - | The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006). | + | The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably [https://en.wikipedia.org/wiki/Histone_H2A H2A histone] mRNA export (Hargous 2006). |
<StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | <StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | ||
| Line 31: | Line 31: | ||
=== SR Domain === | === SR Domain === | ||
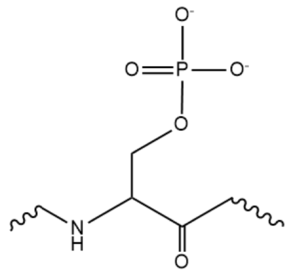
| - | While the RNA-protein interaction occurs at the RRMs, SR domains are typically responsible for the recruitment of other proteins that act in the splicing mechanism. The serines within the SR domain are phosphorylated by kinases within the cell to direct them to pre-mRNA sites (Figure 4). [[Image:phosphorylatedserine.png|300px|right|thumb|'''Figure 4.''' An example of one phosphorylated serine within the RS domain.]] Phosphorylation acts as recruiting tools for the SRp20 protein and other SR proteins to promote splicing. However, some SR proteins have shown that phosphorylation actually leads to a decrease in splicing, as in the SRp38 protein undergoing heat shock. Research has recently revealed that serines are dephosphorylated as splicing continues, indicating how far along the mRNA strand is in splicing. Dephosphorylation then serves as a signal to the cell that the mRNA is ready to be exported out of the nucleus. Rephosphorylation then appears to trigger the SR protein to enter back into the nucleus. | + | While the RNA-protein interaction occurs at the RRMs, SR domains are typically responsible for the recruitment of other proteins that act in the splicing mechanism. The serines within the SR domain are phosphorylated by kinases within the cell to direct them to pre-mRNA sites (Figure 4). [[Image:phosphorylatedserine.png|300px|right|thumb|'''Figure 4.''' An example of one phosphorylated serine within the RS domain.]] Phosphorylation acts as recruiting tools for the SRp20 protein and other SR proteins to promote splicing. However, some SR proteins have shown that phosphorylation actually leads to a decrease in splicing, as in the [https://www.ncbi.nlm.nih.gov/pubmed/14765198 SRp38] protein undergoing heat shock. Research has recently revealed that serines are dephosphorylated as splicing continues, indicating how far along the mRNA strand is in splicing. Dephosphorylation then serves as a signal to the cell that the mRNA is ready to be exported out of the nucleus. Rephosphorylation then appears to trigger the SR protein to enter back into the nucleus. |
The SR domain also appears to modulate mRNA stability, though different SR proteins have been shown to have greater effects. Some SR proteins appears to increase the sensitivity of certain mRNA sequences to degradation; this degradation appears to be controlled by SR domain interactions with the 3’ UTR of the pre-mRNA. These effects are not present without the SR domain of the protein. The destabilizing effect of SRp20 specifically has yet to be studied. (Huang & Steitz 2003). | The SR domain also appears to modulate mRNA stability, though different SR proteins have been shown to have greater effects. Some SR proteins appears to increase the sensitivity of certain mRNA sequences to degradation; this degradation appears to be controlled by SR domain interactions with the 3’ UTR of the pre-mRNA. These effects are not present without the SR domain of the protein. The destabilizing effect of SRp20 specifically has yet to be studied. (Huang & Steitz 2003). | ||
== 9G8 and SRP20 == | == 9G8 and SRP20 == | ||
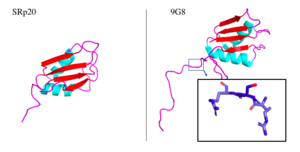
| - | 9G8 is another SR protein that is 80% similar in amino acid sequence. They are two of the smallest proteins in the SR family and both contain an RRM that promotes export of mRNA through interaction with the TAP protein. The only other protein shown to promote transport through the TAP protein is SF2, one of the first SR proteins discovered (Huang 2003). Both RRMs adopt a βαββαβ mentioned earlier. Both RRMs interact with RNA with limited selectivity and therefore recognize many different RNA sequences. The 9G8 RRM contains an large hydrophobic core on its B-sheet. Observing the 9G8 protein has proved useful in understanding SRp20 less stable protein structure. In fact, the one of the only significant structural difference between the two proteins lies in the loops between a-helix 2 and B-4 where several amino acids are not conserved. | + | [https://en.wikipedia.org/wiki/SFRS7 9G8] is another SR protein that is 80% similar in amino acid sequence. They are two of the smallest proteins in the SR family and both contain an RRM that promotes export of mRNA through interaction with the TAP protein. The only other protein shown to promote transport through the TAP protein is SF2, one of the first SR proteins discovered (Huang 2003). Both RRMs adopt a βαββαβ mentioned earlier. Both RRMs interact with RNA with limited selectivity and therefore recognize many different RNA sequences. The 9G8 RRM contains an large hydrophobic core on its B-sheet. Observing the 9G8 protein has proved useful in understanding SRp20 less stable protein structure. In fact, the one of the only significant structural difference between the two proteins lies in the loops between a-helix 2 and B-4 where several amino acids are not conserved. |
Aside from the RRM, both proteins have one SR-rich domain although 9G8 includes about 40 more amino acids this domain (Corbo 2013). [[Image:9G8srp20comparison.png|300px|right|thumb|'''Figure 5.''' SRp20 and 9G8 proteins. The PDB file for SRp20 (left; PDB file: 2i2y) does not include the SR-rich domain but 9G8 (right; PDB file: 2hvz) does. Boxed image shows an RSR region in the PG8 protein with oxygens highlighted in red. Images taken from PyMol software.jpg]] Within the bigger 9G8 protein, there is a zinc knuckle that allows for binding of pyrimidine-rich RNA sequences. This zinc knuckle is not present in SRp20, lending the protein to binding of more purine-rich sequences (Huang 2003).Not only are these two proteins similar, but they also play similar roles in mRNA export out of the nucleus. Both move continuously between the nucleus and cytoplasm which requires phosphorylation of its serine residues located in the SR-rich domain. Serine phosphorylation has been shown to have great importance in the proper functioning of many SR proteins (Figure Shepard & Hertel 2009). | Aside from the RRM, both proteins have one SR-rich domain although 9G8 includes about 40 more amino acids this domain (Corbo 2013). [[Image:9G8srp20comparison.png|300px|right|thumb|'''Figure 5.''' SRp20 and 9G8 proteins. The PDB file for SRp20 (left; PDB file: 2i2y) does not include the SR-rich domain but 9G8 (right; PDB file: 2hvz) does. Boxed image shows an RSR region in the PG8 protein with oxygens highlighted in red. Images taken from PyMol software.jpg]] Within the bigger 9G8 protein, there is a zinc knuckle that allows for binding of pyrimidine-rich RNA sequences. This zinc knuckle is not present in SRp20, lending the protein to binding of more purine-rich sequences (Huang 2003).Not only are these two proteins similar, but they also play similar roles in mRNA export out of the nucleus. Both move continuously between the nucleus and cytoplasm which requires phosphorylation of its serine residues located in the SR-rich domain. Serine phosphorylation has been shown to have great importance in the proper functioning of many SR proteins (Figure Shepard & Hertel 2009). | ||
Revision as of 02:24, 3 April 2018
Contents |
Introduction
Overview
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other eukaryotes. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM)(Corbo, 2013).
History
Splicing is one step in the process of RNA maturation that cuts out introns and joins exons together. Both the spliceosome, a complex of snRNAs (U1, U2, etc.), and splicing factors like SRp20 interact with intron consensus sequences in the pre-mRNA to regulate this process. Alternative splicing allows one mRNA molecule to produce numerous proteins that perform different functions in a cell by inclusion and exclusion of RNA sequences. There are two main families of splicing factors: Serine-Arginine rich (SR) proteins and heterogeneous nuclear RiboNucleoProteins (hnRNPs). The SRp20 protein belongs to the SR protein family. All SR proteins are defined by a RNA-binding domain at the N-terminus and a serine-arginine rich domain at the C-terminus (Corbo et al. 2013). The discovery of this family started in the 1900s with the SF2 (SRp30a) protein and has since come to include twelve proteins, all of which act as splicing factors. SRp20 was first discovered in calf thymus when it was separated with several other SR proteins based on their molecular weight (Zhaler 1992). An identical protein, called X16, was discovered in an earlier paper studying different genes that change expression during B-cell development (Ayane 1991). At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true (Ayane 1991; Corbo et al. 2013). The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease (Corbo 2013). SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4 (Corbo 2013). Another function of SRp20 is its role in export of mRNA out of the nucleus, notably H2A histone mRNA export (Hargous 2006).
| |||||||||||
References
9. Shepard, P. J., & Hertel, K. J. 2009. “The SR protein family.” Genome biology; 10(10):242.