User:Neel Bhagat/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
== Introduction == | == Introduction == | ||
| - | + | <StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | |
=== Overview === | === Overview === | ||
The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM)<ref name="Corbo2013">PMID:23685143</ref>. | The SRp20 protein is an alternative splicing factor found in homo sapiens as well as many other [https://en.wikipedia.org/wiki/Eukaryote eukaryotes]. It is a relatively small protein with a length of 164 amino acids and a weight of about 19kDa. In fact, it is the smallest member of the SR protein family. The protein contains two domains: a serine-arginine rich (SR) domain and a RNA-recognition domain (RRM)<ref name="Corbo2013">PMID:23685143</ref>. | ||
| Line 9: | Line 9: | ||
An identical protein, called [http://www.uniprot.org/uniprot/Q9V3V0 X16], was discovered in an earlier paper studying different genes that change expression during [https://en.wikipedia.org/wiki/B_cell B-cell] development<ref name="Corbo2013">PMID:23685143</ref>. At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true<ref name="Ayane">PMID:2030943</ref><ref name="Cacero">PMID:11932019</ref>. | An identical protein, called [http://www.uniprot.org/uniprot/Q9V3V0 X16], was discovered in an earlier paper studying different genes that change expression during [https://en.wikipedia.org/wiki/B_cell B-cell] development<ref name="Corbo2013">PMID:23685143</ref>. At the time, the protein was assumed to play a role in RNA processing and cellular proliferation, a finding that was later proved to be true<ref name="Ayane">PMID:2030943</ref><ref name="Cacero">PMID:11932019</ref>. | ||
The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease<ref name="Corbo2013">PMID:23685143</ref>. SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4<ref name="Corbo2013">PMID:23685143</ref>. Another function of SRp20 is its role in export of mRNA out of the nucleus, notably [https://en.wikipedia.org/wiki/Histone_H2A H2A histone] mRNA export<ref name="Hargous">PMID:17036044</ref>. | The SRp20 protein has been shown to play a role in cancer progression and neurological disorders, specifically through alternative splicing. For example, SRp20 has been shown to play a role in alternative splicing of the Tau protein, an integral protein in the progression of Alzheimer’s disease<ref name="Corbo2013">PMID:23685143</ref>. SRp20 has even been found to serve as a splicing factor for its own mRNA, influencing the inclusion of exon 4<ref name="Corbo2013">PMID:23685143</ref>. Another function of SRp20 is its role in export of mRNA out of the nucleus, notably [https://en.wikipedia.org/wiki/Histone_H2A H2A histone] mRNA export<ref name="Hargous">PMID:17036044</ref>. | ||
| - | <StructureSection load='2i2y' size='400' side='right' caption='SRp20 bound to RNA ligand and IgG binding domain 1 (PDB entry [[2i2y]])' scene=''> | ||
=== Structure Determination === | === Structure Determination === | ||
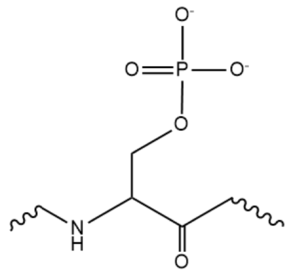
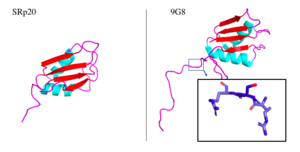
| - | Attempts to determine the structure of native SRp20 have been largely unsuccessful due to the low solubility of the protein. This is likely due to the hydrophobic core of the RRM and exposed hydrophobic residues for RNA recognition on the β-sheets. As a solution, researchers removed the SR domain from the C terminus, leaving only the SRp20 RRM and a small arginine rich segment at the C terminus, then fused with a soluble <scene name='78/782597/Imager0/2'>IgG binding domain</scene> of Streptococcal protein G to the N terminus of the protein, providing the first published structure of the SRp20 RRM via | + | Attempts to determine the structure of native SRp20 have been largely unsuccessful due to the low solubility of the protein. This is likely due to the hydrophobic core of the RRM and exposed hydrophobic residues for RNA recognition on the β-sheets. As a solution, researchers removed the SR domain from the C terminus, leaving only the SRp20 RRM and a small arginine rich segment at the C terminus, then fused with a soluble <scene name='78/782597/Imager0/2'>IgG binding domain</scene> of Streptococcal protein G to the N terminus of the protein, providing the first published structure of the SRp20 RRM via NMR. However, the solution of the structure via [https://en.wikipedia.org/wiki/Nuclear_magnetic_resonance NMR], in addition to fusion with a globular tag, results in multiple possible conformations of the protein, meaning measurements such as bond angles, lengths, and substrate interactions are variable. Further, information concerning structural aspects of the SR domain are still limited to experimental data of protein function with certain mutations or deletions, and by comparison to sister proteins such as 9G8. To date, structure of the SR domain or the protein without the globular tag have not been solved, nor has a crystal structure for any part of the protein been determined<ref name="Hargous">PMID:17036044</ref>. |
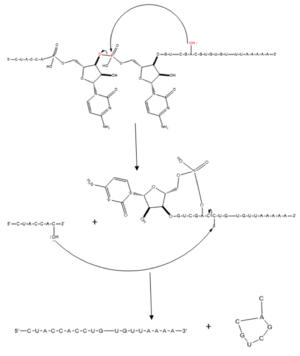
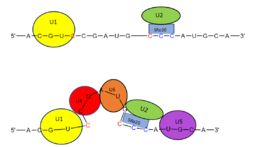
== Splicing Activity == | == Splicing Activity == | ||
| Line 22: | Line 21: | ||
=== RNA Recognition Motif === | === RNA Recognition Motif === | ||
| - | The SRP20 RRM (aa 1-86) a βαββαβ <scene name='78/782597/Imager1/2'>pattern</scene>, common of many other RRMs3. For substrate binding, researchers used a 4 base RNA with sequence CAUC, which matches the SRP20 recognition sequence found on corresponding H2A mRNA. The RNA bases each <scene name='78/782597/Imager2/5'>stack</scene> onto an aromatic side chain protruding from one of the SRP20 β-sheets, forming the primary interactions which allow substrate binding to the protein. In particular, C1 <scene name='78/782597/Imager3/2'>stacks</scene> on Y13 in β1, <scene name='78/782597/Imager4/2'>A2</scene> stacks on F50 in β3, and F48 of β3 sits in between the sugar rings of C1 and A2. It should also be noted that A2 adopts an irregular <scene name='78/782597/Imager5/3'>syn</scene> conformation when bound to the RRM, something that was observed only for guanine in the 2 position | + | The SRP20 RRM (aa 1-86) a βαββαβ <scene name='78/782597/Imager1/2'>pattern</scene>, common of many other RRMs3. For substrate binding, researchers used a 4 base RNA with sequence CAUC, which matches the SRP20 recognition sequence found on corresponding H2A mRNA. The RNA bases each <scene name='78/782597/Imager2/5'>stack</scene> onto an aromatic side chain protruding from one of the SRP20 β-sheets, forming the primary interactions which allow substrate binding to the protein. In particular, C1 <scene name='78/782597/Imager3/2'>stacks</scene> on Y13 in β1, <scene name='78/782597/Imager4/2'>A2</scene> stacks on F50 in β3, and F48 of β3 sits in between the sugar rings of C1 and A2. It should also be noted that A2 adopts an irregular <scene name='78/782597/Imager5/3'>syn</scene> conformation when bound to the RRM, something that was observed only for guanine in the 2 position previously. U3 <scene name='78/782597/Imager8/4'>Stacks</scene> onto F48 in β3, also W40 and A42 in β2, however when bound, U3 <scene name='78/782597/Imager9/2'>bulges</scene> out of line in comparison to the rest of the substrate. C4 partially stacks over <scene name='78/782597/Imager6/4'>A2</scene>, and also forms hydrogen <scene name='78/782597/Imager7/3'>bonds</scene> between the C4 amino group, A2 2’ oxygen, and a main chain phosphate oxygen. |
While all 4 bases form a number of hydrophobic stacking interactions, alteration to the last 3 bases of substrate sequence does not significantly impact binding affinity, while C to G mutation of C1 results in a 10-fold decrease in binding affinity. This suggests that C1 interacts specifically with the protein, while positions 2-4 interact nonspecifically3. The Srp20 RRM is able to recognize C1 with high specificity primarily through 4 <scene name='78/782597/Imager10/3'>hydrogen bonds</scene>: from the C1 amino protons to Leu 80 backbone carbonyl oxygen and to Glu 79 side-chain carbonyl oxygen, from C1 N3 to Asn82 amide, and C1 O2 with Ser 81 side chain hydroxyl group. | While all 4 bases form a number of hydrophobic stacking interactions, alteration to the last 3 bases of substrate sequence does not significantly impact binding affinity, while C to G mutation of C1 results in a 10-fold decrease in binding affinity. This suggests that C1 interacts specifically with the protein, while positions 2-4 interact nonspecifically3. The Srp20 RRM is able to recognize C1 with high specificity primarily through 4 <scene name='78/782597/Imager10/3'>hydrogen bonds</scene>: from the C1 amino protons to Leu 80 backbone carbonyl oxygen and to Glu 79 side-chain carbonyl oxygen, from C1 N3 to Asn82 amide, and C1 O2 with Ser 81 side chain hydroxyl group. | ||
The semi specific RNA recognition is a mechanism which reduces evolutionary pressure on bound mRNA by increasing the number of possible RNA recognition sequences. As a result, tolerance for possible mutation in the RNA sequence is increased, meaning Srp20 can bind a more diverse range of substrates, or even original substrates that were mutated during replication (eg. H2A mRNA with a point mutation) thereby increasing organism survival chance by reducing the probability of physiological impact as a result of certain mutations<ref name="Hargous">PMID:17036044</ref>. | The semi specific RNA recognition is a mechanism which reduces evolutionary pressure on bound mRNA by increasing the number of possible RNA recognition sequences. As a result, tolerance for possible mutation in the RNA sequence is increased, meaning Srp20 can bind a more diverse range of substrates, or even original substrates that were mutated during replication (eg. H2A mRNA with a point mutation) thereby increasing organism survival chance by reducing the probability of physiological impact as a result of certain mutations<ref name="Hargous">PMID:17036044</ref>. | ||
| Line 49: | Line 48: | ||
SRp20 and other SR proteins have been shown to prevent [https://en.wikipedia.org/wiki/R-loop R-loops] from forming, 3-stranded nucleic acid structures consisting of RNA and DNA. R-loops have been known to promote mutations, recombination, and chromosome rearrangement. One proposed mechanism for R-loop prevention by SRp20 is that SRp20, being a protein involved in RNA metabolism, is a binding partner of the [https://www.uniprot.org/uniprot/P11387 TOP1] protein. Underexpression of TOP1 also promotes R-loop formation. TOP1 has kinase activity that potentially phosphorylates the SR domain of SRp20, which contributes to its function. Underexpression of TOP1 would lead to loss-of-function of SRp20, which could lead to cancer and Alzheimers as mentioned above, as well as cause R-loops to form. R-loops have been associated with disorders such as [https://www.ndss.org/about-down-syndrome/down-syndrome/ Down Syndrome]<ref name="Naro">PMID:25926848</ref>. | SRp20 and other SR proteins have been shown to prevent [https://en.wikipedia.org/wiki/R-loop R-loops] from forming, 3-stranded nucleic acid structures consisting of RNA and DNA. R-loops have been known to promote mutations, recombination, and chromosome rearrangement. One proposed mechanism for R-loop prevention by SRp20 is that SRp20, being a protein involved in RNA metabolism, is a binding partner of the [https://www.uniprot.org/uniprot/P11387 TOP1] protein. Underexpression of TOP1 also promotes R-loop formation. TOP1 has kinase activity that potentially phosphorylates the SR domain of SRp20, which contributes to its function. Underexpression of TOP1 would lead to loss-of-function of SRp20, which could lead to cancer and Alzheimers as mentioned above, as well as cause R-loops to form. R-loops have been associated with disorders such as [https://www.ndss.org/about-down-syndrome/down-syndrome/ Down Syndrome]<ref name="Naro">PMID:25926848</ref>. | ||
| - | + | == References == | |
<references/> | <references/> | ||
Revision as of 18:09, 6 April 2018
Introduction
| |||||||||||