We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1449
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
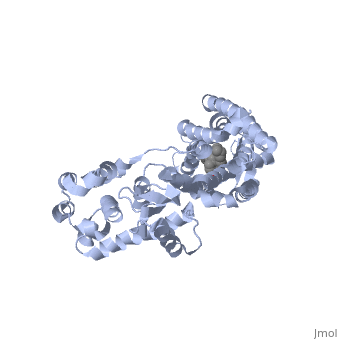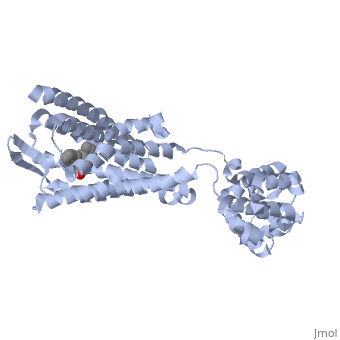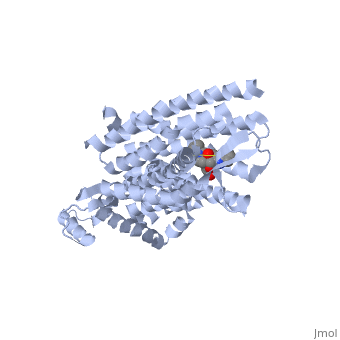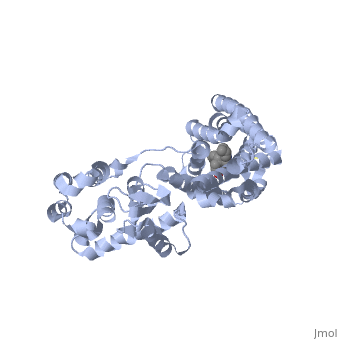
| - | + | Opioid receptors are G-protein coupled receptors (GPCR), which bind endogenous opioid peptide neurotransmitters (such as enkephalins and endorphins) and exogenous synthetic opiate drugs (such as morphine, codeine, and heroin) as ligands to hinder pain-signaling in the brain, peripheral nerves, and digestive tract. μ-opioid receptors are one of the four major classes of opioid receptors, which also includes δ-opioid receptors, κ-opioid receptors, and nociceptin opioid receptors. The μ-opioid receptor MOR-1 is expressed by the gene OPRM1 in vertebrates. The molecular structure of MOR-1 was better understood after its cloning in 1993. According to the American Society for Pharmacology and Experimental Therapeutics, the amino acid sequence of MOR-1 is 60-70% homologous to the other classes of opioid receptors. The difference between MOR-1 and the other opioid receptor proteins lies in its extracellular N-terminus, intracellular C-terminus, and second and third extracellular loops. The μ-opioid receptor is a 7-multispanning integral membrane protein found in dorsal root ganglion cells and peripheral nerve cells in humans, with its binding site exposed to the extracellular surface. The transmembrane domain of MOR-1 will dimerize at TM5 and TM6 to form oligomers. MOR-1 has important implications as a target for pain relievers as well as a treatment for drug abuse. | |
== Function == | == Function == | ||
| Line 23: | Line 23: | ||
<references/> | <references/> | ||
| - | http:// | + | 1. Pan, G. W. (2013, October 01). Mu Opioids and Their Receptors: Evolution of a Concept. Retrieved April 11, 2018, from http://pharmrev.aspetjournals.org/content/65/4/1257 |
| + | |||
| + | 2. Kaserer, T., Lantero, A., Schmidhammer, H., Spetea, M., & Schuster, D. (2016, February 18). μ Opioid receptor: Novel antagonists and structural modeling. Retrieved April 11, 2018, from https://www.nature.com/articles/srep21548 | ||
Revision as of 19:44, 11 April 2018
| This Sandbox is Reserved from Jan 22 through May 22, 2018 for use in the course Biochemistry II taught by Jason Telford at the Maryville University, St. Louis, Missouri, USA. This reservation includes Sandbox Reserved 1446 through Sandbox Reserved 1455. |
To get started:
More help: Help:Editing |
Delta Opioid Receptor
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
1. Pan, G. W. (2013, October 01). Mu Opioids and Their Receptors: Evolution of a Concept. Retrieved April 11, 2018, from http://pharmrev.aspetjournals.org/content/65/4/1257
2. Kaserer, T., Lantero, A., Schmidhammer, H., Spetea, M., & Schuster, D. (2016, February 18). μ Opioid receptor: Novel antagonists and structural modeling. Retrieved April 11, 2018, from https://www.nature.com/articles/srep21548