We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Poly(A) binding protein
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
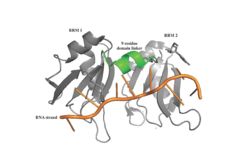
[[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | [[Image: PABP Biological Assembly 1.jpg |250 px|left|thumb|Figure 1: PABP Biological Assembly with linker highlighted. ]] | ||
| - | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the [https://en.wikipedia.org/wiki/Polyadenylation 3' poly(A) tail] of [https://en.wikipedia.org/wiki/Messenger_RNA mRNA] that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and [https://en.wikipedia.org/wiki/Initiation_factor initiation factors] causes it to also play a significant role in [https://en.wikipedia.org/wiki/Eukaryotic_translation translation] initiation and mRNA stabilization and degradation. PABP consists of four conserved [https://en.wikipedia.org/wiki/Protein_domain domains] of [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motifs] (RRMs); however, the two [https://en.wikipedia.org/wiki/N-terminus N-terminal] RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of full-length PABP, so due to this and the modular organization of the protein, it is thought that the full complement of PABP may not be essential. Thus, the published [https://en.wikipedia.org/wiki/X-ray_scattering_techniques X-ray diffraction] structure | + | Human Poly(A) Binding Protein (PABP) is a biopolypeptide involved in recognizing the [https://en.wikipedia.org/wiki/Polyadenylation 3' poly(A) tail] of [https://en.wikipedia.org/wiki/Messenger_RNA mRNA] that is added to an mRNA transcript during mRNA processing.This recognition as well as PABP's interaction with other proteins and [https://en.wikipedia.org/wiki/Initiation_factor initiation factors] causes it to also play a significant role in [https://en.wikipedia.org/wiki/Eukaryotic_translation translation] initiation and mRNA stabilization and degradation. PABP consists of four conserved [https://en.wikipedia.org/wiki/Protein_domain domains] of [https://en.wikipedia.org/wiki/RNA_recognition_motif RNA recognition motifs] (RRMs); however, the two [https://en.wikipedia.org/wiki/N-terminus N-terminal] RRMs (RRM1 and RRM2) and the short linker sequence that connects them supports most of the function of full-length PABP, so due to this and the modular organization of the protein, it is thought that the full complement of PABP may not be essential. Thus, the published [https://en.wikipedia.org/wiki/X-ray_scattering_techniques X-ray diffraction] structure reveals RRM1 and RRM2 at a 2.6 Angstrom [https://en.wikipedia.org/wiki/Resolution_(electron_density) resolution]. This is shown as <scene name='78/781947/Biological_assembly_1/1'>Biological Assembly 1</scene>. Both RRM 1 and 2 are needed to support biochemical function, that is, no one RRM can support biochemical function. Additionally, there is a [https://en.wikipedia.org/wiki/Proline proline] rich [https://en.wikipedia.org/wiki/C-terminus C-terminal] portion of variable length that is not well conserved and unknown as to how it contributes to the protein's function <ref name= "PABP"> Deo, Rahul C, et al. “Recognition of Polyadenylate RNA by the Poly(A)-Binding Protein.” Cell, vol. 98, no. 6, 1999, pp. 835–845., doi:10.1016/s0092-8674(00)81517-2.</ref>. |
== Function and Structure == | == Function and Structure == | ||
| Line 21: | Line 21: | ||
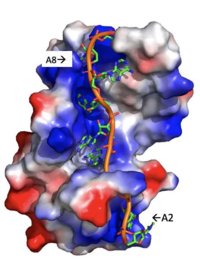
====Adenosine Stabilization Interaction Patterns==== | ====Adenosine Stabilization Interaction Patterns==== | ||
| - | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781947/Interactions_with_a2/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between [https://en.wikipedia.org/wiki/Aromatic_amino_acid aromatic] and [https://en.wikipedia.org/wiki/Aliphatic_compound alipathic] side chains. Stacking interactions also occur with adenines between aromatic and alipathic side chains, such as <scene name='78/781947/Stacking_interactions_a3/1'>Adenosine-3 stacking interactions</scene> occur between Phe 102 and Arg 179, and it is specified by [https://en.wikipedia.org/wiki/Lysine Lysine] 104. Additionally, <scene name='78/781947/Residues_interacting_with_a6/ | + | Specifically, there are several significant interaction patterns that stabilize adenosine recognition. RRM 1 and 2 makes significant interactions with the adenosine backbone, shown in Figure 3. Additionally, the adenosine stabilizes itself within the binding by intramolecular stacking interactions between adenosines. Through the extensive <scene name='78/781947/Interactions_with_a2/1'>interactions with adenosine 2</scene>, the RRM specifies the position of adenosine 2, allowing it to make strong intramolecular stacking interactions with adenosine 1. As a result, adenosine 1 requires less contact with the RRM, as it is mostly stabilized by adenosine 2. Furthermore, some adenosines like adenosine 3 and adenosine 6 are stabilized by being sandwiched between [https://en.wikipedia.org/wiki/Aromatic_amino_acid aromatic] and [https://en.wikipedia.org/wiki/Aliphatic_compound alipathic] side chains. Stacking interactions also occur with adenines between aromatic and alipathic side chains, such as <scene name='78/781947/Stacking_interactions_a3/1'>Adenosine-3 stacking interactions</scene> occur between Phe 102 and Arg 179, and it is specified by [https://en.wikipedia.org/wiki/Lysine Lysine] 104. Additionally, <scene name='78/781947/Residues_interacting_with_a6/3'>Adenosine-6 stacking interactions</scene> occurs similarly between Tyr 14 and Arg 94, but it is specified doubly by two residues, Trp-86 and Gln-88 <ref name= "PABP"/>. |
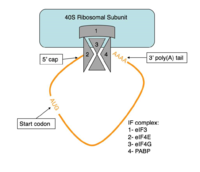
===Translation Initiation=== | ===Translation Initiation=== | ||
| Line 37: | Line 37: | ||
====Structural Components of PABP Translation Initiation==== | ====Structural Components of PABP Translation Initiation==== | ||
| - | The RRMs support interactions with the interacting proteins such as eIF4G and [https://en.wikipedia.org/wiki/PAIP1 PAIP-1], however, the specific ways in which PABP interacts with these proteins are not structurally proven. However, there is a convex dorsal surface present on the RRM 1 and 2 motifs formed by the two α-helices in each RRM, specified as H1 and H2 in RRM1 and H1' and H2' in RRM2. This surface contains a sequence portion of <scene name='78/781947/H1_and_h2_h2ophobic_residues/4'>conserved hydrophobic residues</scene> and <scene name='78/781947/Hydrophilic_residues/ | + | The RRMs support interactions with the interacting proteins such as eIF4G and [https://en.wikipedia.org/wiki/PAIP1 PAIP-1], however, the specific ways in which PABP interacts with these proteins are not structurally proven. However, there is a convex dorsal surface present on the RRM 1 and 2 motifs formed by the two α-helices in each RRM, specified as H1 and H2 in RRM1 and H1' and H2' in RRM2. This surface contains a sequence portion of <scene name='78/781947/H1_and_h2_h2ophobic_residues/4'>conserved hydrophobic residues</scene> and <scene name='78/781947/Hydrophilic_residues/3'>conserved hydrophilic residues</scene> |
residues. It is thought that this area of conservation thus produces overlapping binding sites to interact with eIF4G and PAIP-1. For example, the conserved acidic residues may be beneficial to be used in to interact with essential basic residues present in both eIF4G and PAIP-1 via ionic interactions <ref name="PABP"/>. While there are only proposed mechanisms for how PABP promotes the initiation of translation in Homo sapiens, there is a pathogenic [https://en.wikipedia.org/wiki/Protozoa protozoan], Leishmania, that through a study done by Osvaldo P. de Melo Neto et. al, a site on the domain linker of a PABP homolog, PABP-1, of either serine-proline or threonine-proline residues which an eIF4F-like complex [https://en.wikipedia.org/wiki/Phosphorylation phosphorylates]. The authors suggest that this phosphorylation is part of how PABP-1 aids the eIF4F complex in initiating translation. They supported this by removing the gene that encodes PABP-1 and the results showed that the protozoan could not initiated cell growth and therefore survive without the PABP-1 gene. The homo sapiens' PABP also contain a (<scene name='78/781947/Pro-ser_in_linker/2'>Serine-Proline</scene>) site on the domain linker which could be interacting with the eIF4 complex in a similar way as in Leishmania protozoan <ref name="Osvaldo">De Melo Neto, Osvaldo P., et al. “Phosphorylation and Interactions Associated with the Control of the Leishmania Poly-A Binding Protein 1 (PABP1) Function during Translation Initiation.” RNA Biology, 23 Mar. 2018, pp. 1–17., doi:10.1080/15476286.2018.1445958.</ref>. | residues. It is thought that this area of conservation thus produces overlapping binding sites to interact with eIF4G and PAIP-1. For example, the conserved acidic residues may be beneficial to be used in to interact with essential basic residues present in both eIF4G and PAIP-1 via ionic interactions <ref name="PABP"/>. While there are only proposed mechanisms for how PABP promotes the initiation of translation in Homo sapiens, there is a pathogenic [https://en.wikipedia.org/wiki/Protozoa protozoan], Leishmania, that through a study done by Osvaldo P. de Melo Neto et. al, a site on the domain linker of a PABP homolog, PABP-1, of either serine-proline or threonine-proline residues which an eIF4F-like complex [https://en.wikipedia.org/wiki/Phosphorylation phosphorylates]. The authors suggest that this phosphorylation is part of how PABP-1 aids the eIF4F complex in initiating translation. They supported this by removing the gene that encodes PABP-1 and the results showed that the protozoan could not initiated cell growth and therefore survive without the PABP-1 gene. The homo sapiens' PABP also contain a (<scene name='78/781947/Pro-ser_in_linker/2'>Serine-Proline</scene>) site on the domain linker which could be interacting with the eIF4 complex in a similar way as in Leishmania protozoan <ref name="Osvaldo">De Melo Neto, Osvaldo P., et al. “Phosphorylation and Interactions Associated with the Control of the Leishmania Poly-A Binding Protein 1 (PABP1) Function during Translation Initiation.” RNA Biology, 23 Mar. 2018, pp. 1–17., doi:10.1080/15476286.2018.1445958.</ref>. | ||
Revision as of 16:48, 17 April 2018
Poly(A) binding protein
| |||||||||||