This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Khadar Abdi/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Threonyl-tRNA Synthetase/ligase== | ==Threonyl-tRNA Synthetase/ligase== | ||
| - | <StructureSection load='1NYQ' size=' | + | <StructureSection load='1NYQ' size='300' side='right' caption='Staphylococcus aureus threonyl-tRNA Synthetase bound to Threonyl-Sulfamoyl Adenosine' scene=''> |
== Introduction == | == Introduction == | ||
| Line 17: | Line 17: | ||
== Structural highlights== | == Structural highlights== | ||
| - | As mentioned earlier, TARS is homodimer protein. The protein is mainly classified as a alpha beta protein | + | As mentioned earlier, TARS is homodimer protein found in two forms of a cell, mitochondrial and cytoplasmic. Most of the structures seen will primarly be cytoplasmic. The protein is mainly classified as a alpha beta protein in both bacteria and eukaryotic cells<ref>PMID:23362938</ref>. Each chain is composed of 4 domain classified by SCOP: a N1 domain, a N2 domain, a catalytic domain and an anti-codon domain<ref>PMID:12875846</ref>.<Structure load='Image of ThrS Staph colored label by domains' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> |
| + | |||
| + | ===N-terminus (Residue 1-241)=== | ||
| + | The N1-Domain and N-2 domain is the found in the N-terminus of the TARS enzyme chain. Specific N1 is found from 1-62 residue while N2 domain is found from 62-241 residue(PDB: [[1TJE]]). Although located in neighboring regions, the N1-domain and N2-domain deviate dramatically from one another. Structurally, N1-domain has a TGS-fold (short for TARS, GTPase, Spo1 domain as their the most common proteins that present this fold)<ref>PMID:10447505</ref> which has 3 antiparallel beta sheets interacting with an alpha helix causing a formation of roll-like structure (similar to the description by Cath: Ubiquitin-like roll)<ref>http://www.rcsb.org/pdb/explore/macroMoleculeData.do?structureId=1TJE</ref>. The structure has no known function but it is pronounced structure found common within TARS. | ||
| + | |||
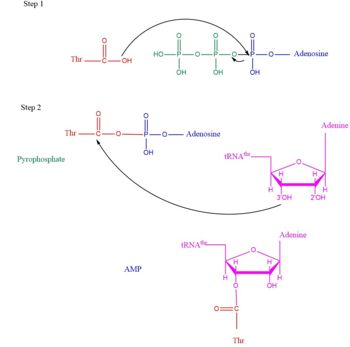
| + | This is the opposite of the N2-domain, or sometimes called TARS additional domain. This is ironic as this addition is very important for proofreading TARS activity<ref>PMID:15525511</ref>. TARS N2-domain is described as 2-layer alpha-beta sandwich comprised of mostly alpha helices. Interesting enough, the superfamily of this motif is found to be TARS and Alanyl-tRNA synthetase common domain. This domain function similar by discriminating Threonine and Alanine from Serine amino acid (Ser) as serine is similar structure to both amino acids. The editing domain functions first by moving Ser bound tRNA-Thr from the catalytic domain to the editing site by breaking the bonds nucleotide A73, C74, and C75 from the catalytic site allowing the acceptor arm of tRNA-Thr to flip to the editing domain of S12-S13 site. Research on editing hydrolysis propose that residue Tyr 103 plays an important role of guiding acceptor arm towards the editing domain. The Ser bound to tRNA is hydrolzed by a water molecule, interacting with His73, acting as a nucleophile to the alpha carbon of serine, follow by protonation by a 2nd water molecule interacting with carbonyl of Met181 and amide side chain Lys156. The fidility mechanism of AA-tRNA binding is much similar to alanine tRNA-ligase found in the C-terminus as they share 40% similarity in residues. The subtle difference of the isoforms mitochondrial TARS from cytoplasmic TARS appears in editing as the mitochondrial TARS doesn't have N2-domain and requires interaction to hydrolyze serine <ref>PMID:22773845</ref>. | ||
| + | |||
| + | ===Catalytic Domain (Residue 243-=== | ||
| + | |||
| + | ===Anti-Codon Domain=== | ||
Revision as of 04:12, 1 May 2018
Threonyl-tRNA Synthetase/ligase
| |||||||||||