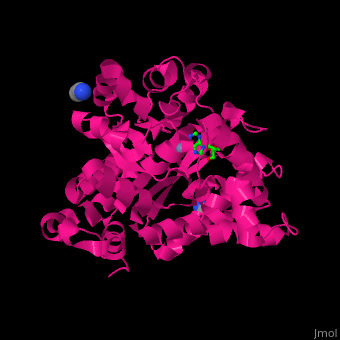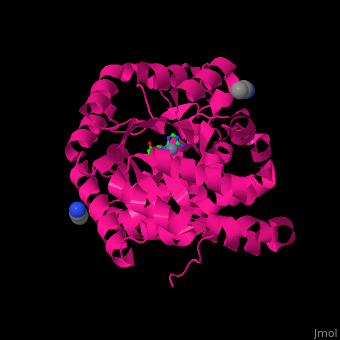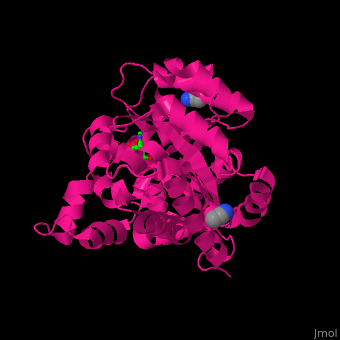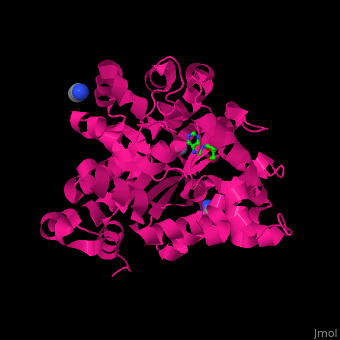This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Adenosine deaminase
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
There are 2 isoforms of ADA: ADA1 and ADA2. <scene name='54/547081/Cv/3'>ADA contains Zn+2 ion in its active site</scene> ([[2pgf]]).<ref>PMID:18602399</ref> Water mlecules are shown as red spheres. | There are 2 isoforms of ADA: ADA1 and ADA2. <scene name='54/547081/Cv/3'>ADA contains Zn+2 ion in its active site</scene> ([[2pgf]]).<ref>PMID:18602399</ref> Water mlecules are shown as red spheres. | ||
| + | |||
| + | == 3D Structures of adenosine deaminase == | ||
| + | [[Adenosine deaminase 3D structures]] | ||
| + | |||
</StructureSection> | </StructureSection> | ||
| Line 57: | Line 61: | ||
**[[3pbm]] – PaADA + Zn + chloropurine <BR /> | **[[3pbm]] – PaADA + Zn + chloropurine <BR /> | ||
**[[3lgg]] – hADA2 + Zn + coformycin<BR /> | **[[3lgg]] – hADA2 + Zn + coformycin<BR /> | ||
| + | **[[6n9m]] – ADA + Zn + pentostatin – ''Salmonella typhimurium''<BR /> | ||
| + | **[[6n91]] – ADA + Zn + pentostatin – ''Vibrio cholerae''<BR /> | ||
*Adenosine deaminase complex with protein | *Adenosine deaminase complex with protein | ||
| Line 63: | Line 69: | ||
**[[2bgn]] – bADA + Zn + dipeptidyl peptidase IV + HIV1 TAT protein peptide<BR /> | **[[2bgn]] – bADA + Zn + dipeptidyl peptidase IV + HIV1 TAT protein peptide<BR /> | ||
| - | *Double-stranded RNA-specific adenosine deaminase | + | *Double-stranded RNA-specific adenosine deaminase; domains – Zα 125-201; Zβ 294-366; 3rd RNA-binding 708-801; catalytic 299-729 |
**[[1xmk]] – hADA Zα domain <br /> | **[[1xmk]] – hADA Zα domain <br /> | ||
**[[1qgp]] – hADA Zα domain - NMR<br /> | **[[1qgp]] – hADA Zα domain - NMR<br /> | ||
**[[2l54]] – hADA Zα domain (mutant) - NMR<br /> | **[[2l54]] – hADA Zα domain (mutant) - NMR<br /> | ||
| + | **[[2acj]], [[1qbj]], [[2gxb]], [[3f21]], [[3f22]], [[3f23]], [[3irq]], [[3irr]], [[5zu1]], [[5zuo]], [[5zup]] – hADA Zα domain + DNA<br /> | ||
**[[2mdr]] – hADA 3rd RNA-binding domain - NMR<br /> | **[[2mdr]] – hADA 3rd RNA-binding domain - NMR<br /> | ||
| - | **[[1zy7]] – hADA + inositol hexakisphosphate<br /> | + | **[[1zy7]] – hADA catalytic domain + inositol hexakisphosphate<br /> |
| - | **[[ | + | **[[6d06]], [[5hp2]], [[5hp3]], [[5ed1]], [[5ed2]] – hADA catalytic domain + RNA<br /> |
| - | **[[2b7t]], [[2b7v]] – rADA – rat<br /> | + | **[[2b7t]], [[2b7v]] – rADA 1st RNA-binding domain – rat<br /> |
| - | **[[2l3c]], [[2l3j]] – rADA + RNA – NMR<br /> | + | **[[2l3c]], [[2l3j]] – rADA 1st RNA-binding domain + RNA – NMR<br /> |
| - | **[[2l2k]] – mADA + RNA – NMR<br /> | + | **[[2l2k]] – mADA residues 230-301 + RNA – NMR<br /> |
**[[2ljh]] – ADA DRBM domain – ''Drosophila melanogaster'' – NMR<br /> | **[[2ljh]] – ADA DRBM domain – ''Drosophila melanogaster'' – NMR<br /> | ||
Revision as of 10:41, 28 February 2019
| |||||||||||
3D Structures of adenosine deaminase
Updated on 28-February-2019
References
- ↑ Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991 May 31;252(5010):1278-84. PMID:1925539
- ↑ Larson ET, Deng W, Krumm BE, Napuli A, Mueller N, Van Voorhis WC, Buckner FS, Fan E, Lauricella A, DeTitta G, Luft J, Zucker F, Hol WG, Verlinde CL, Merritt EA. Structures of substrate- and inhibitor-bound adenosine deaminase from a human malaria parasite show a dramatic conformational change and shed light on drug selectivity. J Mol Biol. 2008 Sep 12;381(4):975-88. Epub 2008 Jun 24. PMID:18602399 doi:http://dx.doi.org/10.1016/j.jmb.2008.06.048