This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jennifer Taylor/Sandbox 5
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
==Structural Highlights== | ==Structural Highlights== | ||
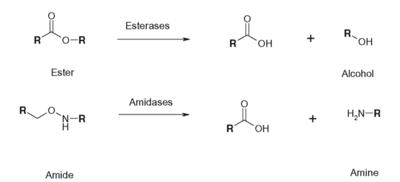
| - | In our research to find similar proven protein structures to our 2QRU because we know that structure dictates function, we used several computational analysis systems to align our sequences and also align our structures. After obtaining the protein, BLAST, PDB, ProMol were used to find the most structurally similar proteins to 2QRU. First, proteins with similar sequences were searched for, because we assumed that if the sequences were similar, than the structures might be as well. On these databases, 1TAH, 1C4X and 3FAK, all in the esterase sub-family, proved to be the most structurally similar after searching for the the motifs and aligned active sites on ProMol. ProMol was important because we were able to find the <scene name='78/787196/2qru_active_site/3'>Active Site of 2QRU</scene>. The active site is where reactions happen, so if the active sites were similar, then maybe the reactions would be so too. We used PyMol to align the 3D structures and the active sites to analyze the similarities between the three proteins with known functions. PyMol allowed us to come to the conclusion that since the three proteins were all in the Esterase family, we could then use an assay that would wither prove or disprove if 2QRU was an esterase. | + | In our research to find similar proven protein structures to our 2QRU because we know that structure dictates function, we used several computational analysis systems to align our sequences and also align our structures. After obtaining the protein, BLAST, PDB, ProMol were used to find the most structurally similar proteins to 2QRU. First, proteins with similar sequences were searched for, because we assumed that if the sequences were similar, than the structures might be as well. On these databases, 1TAH[https://www.rcsb.org/structure/1TAH], 1C4X and [https://www.rcsb.org/structure/3FAK 3FAK], all in the esterase sub-family, proved to be the most structurally similar after searching for the the motifs and aligned active sites on ProMol. ProMol was important because we were able to find the <scene name='78/787196/2qru_active_site/3'>Active Site of 2QRU</scene>. The active site is where reactions happen, so if the active sites were similar, then maybe the reactions would be so too. We used PyMol to align the 3D structures and the active sites to analyze the similarities between the three proteins with known functions. PyMol allowed us to come to the conclusion that since the three proteins were all in the Esterase family, we could then use an assay that would wither prove or disprove if 2QRU was an esterase. |
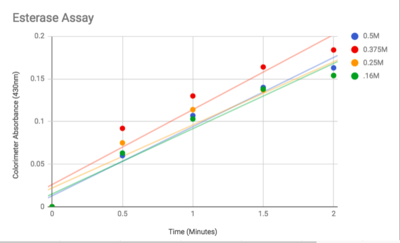
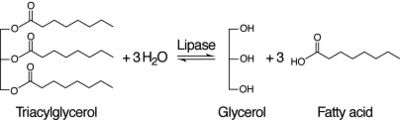
==Functional Assay== | ==Functional Assay== | ||
| Line 38: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| + | 1. Nam K.H., Kim M.Y., Kim S.J., Priyadarshi A., Lee W.H., Hwang K.Y. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. | ||
| + | (2009) Biochem Biophys Res Commun 379: 553-6 | ||
| + | Pubmed Article: 19116143 | ||
| + | |||
<references/> | <references/> | ||
Revision as of 14:37, 21 May 2018
2QRU
Here is a cartoon image of my protein:
| |||||||||||
References
1. Nam K.H., Kim M.Y., Kim S.J., Priyadarshi A., Lee W.H., Hwang K.Y. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. (2009) Biochem Biophys Res Commun 379: 553-6 Pubmed Article: 19116143
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644