We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jennifer Taylor/Sandbox 4
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
== ''In silico'' Analysis == | == ''In silico'' Analysis == | ||
| - | We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. Our top hit was 4M8K, a GDSL-like lipase. | + | [[Image:BLAST_results.png|thumb|left|250px|Figure insert figure#: BLAST results for 4Q7Q]] We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. Our top hit was 4M8K, a GDSL-like lipase. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase. |
Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q was Ser164, Asp193, and His196. We believe that this group of amino acids may be involved in active site of 4Q7Q and therefore affects how the protein works. As seen in this <scene name='78/787192/4q7q_active_site/8'>image</scene>, all three amino acids are close in proximity to one another and are brought together in a single orientation. | Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q was Ser164, Asp193, and His196. We believe that this group of amino acids may be involved in active site of 4Q7Q and therefore affects how the protein works. As seen in this <scene name='78/787192/4q7q_active_site/8'>image</scene>, all three amino acids are close in proximity to one another and are brought together in a single orientation. | ||
| Line 18: | Line 18: | ||
We also performed further analysis in PyMOL and ProMOL which involved the homology of active sites. Top hits included 3LIP, a lipase found in ''Burkholderia cepacia'', 1TAH, a lipase found in ''Burkholderia glumae'', and 1BWR, a hydrolase found in ''Bos taurus''. We aligned putative catalytic triad of 4Q7Q with each of the catalytic triads of these known proteins. | We also performed further analysis in PyMOL and ProMOL which involved the homology of active sites. Top hits included 3LIP, a lipase found in ''Burkholderia cepacia'', 1TAH, a lipase found in ''Burkholderia glumae'', and 1BWR, a hydrolase found in ''Bos taurus''. We aligned putative catalytic triad of 4Q7Q with each of the catalytic triads of these known proteins. | ||
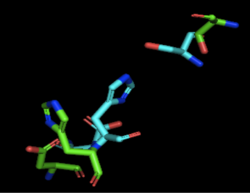
| - | + | [[Image:4Q7Q_3LIP_alignment.png|thumb|left|250px|Figure 1: Alignment of 4Q7Q's putative catalytic triad and 3LIP's catalytic triad: Asp264, Ser87, His286. 4Q7Q is shown in green and 3LIP is shown in blue. The RMS is 2.257.]] 3LIP is a two chained protein. When aligning the catalytic triad of 3LIP using the Catalytic Site Atlas to the putative catalytic triad of 4Q7Q, the RMS is 2.257. | |
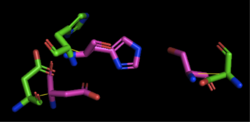
| - | + | [[Image:4Q7Q_1TAH_alignment.png|thumb|right|250px|Figure 2: Alignment of 4Q7Q's putative catalytic triad and 1TAH's catalytic triad: Asp263, Ser87, His285. 4Q7Q is shown in green and 1TAH is shown in pink. The RMS is 2.205.]] 1TAH has four chains. When aligning the catalytic triad of 1TAH to the putative catalytic triad of 4Q7Q, the RMS is 2.205. | |
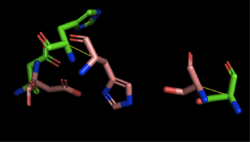
| - | + | [[Image:4Q7Q_1BWR_alignment.png|thumb|left|250px|Figure 3: Alignment of 4Q7Q's putative catalytic triad and 1BWR's catalytic triad: Asp192, Ser47, His195. 4Q7Q is shown in green and 1BWR is shown in pink. The RMS is 2.049.]] 1BWR has one chain. When aligning the catalytic triad of 1BWR to the putative catalytic triad of 4Q7Q, the RMS is 2.049. | |
Compiling all of the data together, we can see that 1BWR is most structurally similar to 4Q7Q due to the low RMS values calculated when aligning both protein structures and catalytic triads. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. | Compiling all of the data together, we can see that 1BWR is most structurally similar to 4Q7Q due to the low RMS values calculated when aligning both protein structures and catalytic triads. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. | ||
Revision as of 20:34, 22 May 2018
4Q7Q
| |||||||||||