4Q7Q is a protein found in Chitinophaga pinensis, a soil bacterium in the sphingobacterial family. Its structure has been previously characterized and exists in Protein Data Bank. Its function, however, has not.
After structural and sequential analysis via various databases including BLAST, Pfam, Dali, PyMOL, and ProMOL, we initially predicted that 4Q7Q is a hydrolase. More specifically, we hypothesized that 4Q7Q is a lipase, an enzyme that can hydrolyze lipids to form fatty acids and a glycerol molecule.
Characteristics of 4Q7Q
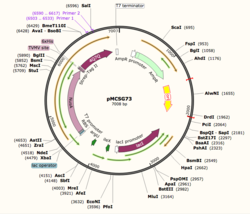
Figure 1: Plasmid Map of pMCSG73, the expression vector for 4Q7Q.
4Q7Q is composed of two chains; one chain can be seen . The colors indicate the translation direction of the peptide sequence from the N to C terminus; red represents the N-terminus while dark blue represents the C-terminus. Based on this structural model, we can see that 4Q7Q is an alpha-beta superfold; there are beta sheets (represented by the straighter strands) sandwiched between the alpha helices (represented by the coiled strands).
Through SnapGene, we analyzed the plasmid pMCSG73, which is the expression vector used to produce 4Q7Q. We found that our plasmid's open reading frame (ORF), the sequence of DNA that will be eventually translated, is 2237 base pairs in length. The size of our protein is 87.1 kDa.
Overall, our ORF is composed of NusA (a transcription elongation factor, two tags- 6XHis and Strep-Tag II, and a TVMV and TEV site. The plasmid map indicates the presence of a lac operon composed of three genes (lacZ, lacY, and lacA), a promoter, terminator, regulator (which codes for a repressor protein), and operator.
In silico Analysis
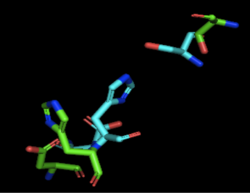
Figure 2: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 3LIP's catalytic triad (shown in blue). The RMS is 2.257.
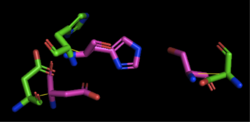
Figure 3: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1TAH's catalytic triad (shown in pink). The RMS is 2.205.
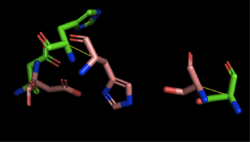
Figure 4: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1BWR's catalytic triad (shown in pink). The RMS is 2.049.
We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. Our top hit was 4M8K, a GDSL-like lipase. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase.
Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q was Ser164, Asp193, and His196. We believe that this group of amino acids may be involved in active site of 4Q7Q and therefore affects how the protein works. As seen in this , all three amino acids are close in proximity to one another and are brought together in a single orientation.
We also performed further analysis in PyMOL and ProMOL which involved the homology of active sites. Top hits included 3LIP, a lipase found in Burkholderia cepacia, 1TAH, a lipase found in Burkholderia glumae, and 1BWR, a hydrolase found in Bos taurus. We aligned putative catalytic triad of 4Q7Q with each of the catalytic triads of these known proteins.
3LIP is a two chained protein. When aligning the catalytic triad of 3LIP (Asp264, Ser87, His286) to the putative catalytic triad of 4Q7Q, the RMS is 2.257.
1TAH has four chains. When aligning the catalytic triad of 1TAH (Asp263, Ser87, His285) to the putative catalytic triad of 4Q7Q, the RMS is 2.205.
1BWR has one chain. When aligning the catalytic triad of 1BWR (Asp192, Ser47, His195) to the putative catalytic triad of 4Q7Q, the RMS is 2.049.
Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the low RMS value calculated. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase.
Bacterial Transformation and Plasmid Purification
Before characterizing the function of 4Q7Q, we first needed to synthesize the protein through first transcribing 4Q7Q's DNA to amplify it and then translating it to express it. First, 4Q7Q's DNA was transcribed using its expression vector, the plasmid pMCS573. Since transformation must occur within a cell, the plasmid was transformed into DH5α cells using protocol from New England Biolabs.
After transformation, DH5α cells were lysed and spread on plates containing LB and ampicillin. Since 4Q7Q's plasmid is ampicillin resistant, only the bacteria that have been transformed with 4Q7Q's plasmid will grow on the plates. 4Q7Q's plasmid was then purified using Zyppy Plasmid Miniprep Kit.
However, although DH5α cells maximize the efficiency of transformations, they do not contain T7 polymerase, which is essential for protein expression. Therefore, the purified plasmid underwent another bacterial transformation into BL21 (DE3) cells that do contain T7 polymerase using protocol from New England Biolabs.
Protein Expression
After the second bacterial transformation, BL21 cells were lysed and spread on LB+amp plates. A bacterial colony was then selected from a plate and suspended in liquid culture. After incubation overnight, the OD260, which essentially measures the concentration of the plasmid DNA, was measured with a biophotometer.
Our plasmid concentration was low, 28.7 (μg/ mL), probably due to the large size of our ORF.
The sample was then induced with IPTG. Essentially, IPTG is a reagent that prevents the repressor from binding to the lac operator to allow expression to occur.
Protein Purification
pNPB Lipase Assay




