This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jennifer Taylor/Sandbox 4
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
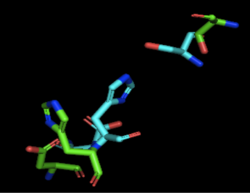
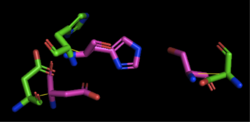
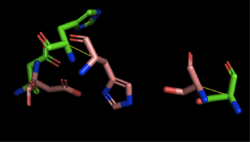
[[Image:4Q7Q_3LIP_alignment.png|thumb|right|250px|Figure 2: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 3LIP's catalytic triad (shown in blue). The RMS is 2.257.]][[Image:4Q7Q_1TAH_alignment.png|thumb|right|250px|Figure 3: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1TAH's catalytic triad (shown in pink). The RMS is 2.205.]][[Image:4Q7Q_1BWR_alignment.png|thumb|right|250px|Figure 4: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1BWR's catalytic triad (shown in pink). The RMS is 2.049.]] | [[Image:4Q7Q_3LIP_alignment.png|thumb|right|250px|Figure 2: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 3LIP's catalytic triad (shown in blue). The RMS is 2.257.]][[Image:4Q7Q_1TAH_alignment.png|thumb|right|250px|Figure 3: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1TAH's catalytic triad (shown in pink). The RMS is 2.205.]][[Image:4Q7Q_1BWR_alignment.png|thumb|right|250px|Figure 4: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1BWR's catalytic triad (shown in pink). The RMS is 2.049.]] | ||
| - | We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. Our top hit was 4M8K, a GDSL-like lipase. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase. | + | We initially analyzed 4Q7Q through the protein structure databases BLAST, Pfam, and Dali. Our top hit was 4M8K, a GDSL-like lipase, a type of a lipase that possesses a flexible active site and therefore has broad substrate specificity. Through BLAST, we found that 4M8K and 4Q7Q had a 36% sequence identity, with an E value of 0.002, indicating that it is a significant match. Since we can use the principle of homology to predict the function of an unknown protein, we first hypothesized that 4Q7Q was too a lipase. |
Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q was Ser164, Asp193, and His196. We believe that this group of amino acids may be involved in the active site of 4Q7Q and therefore affects how the protein works. As seen in this <scene name='78/787192/4q7q_active_site/8'>image</scene>, all three amino acids are close in proximity to one another and are brought together in a single orientation. | Through analyzing the sequence of 4Q7Q in SnapGene and then analyzing the 3D structure in PyMOL, we hypothesized that a possible catalytic triad of 4Q7Q was Ser164, Asp193, and His196. We believe that this group of amino acids may be involved in the active site of 4Q7Q and therefore affects how the protein works. As seen in this <scene name='78/787192/4q7q_active_site/8'>image</scene>, all three amino acids are close in proximity to one another and are brought together in a single orientation. | ||
| Line 34: | Line 34: | ||
1BWR has one chain. When aligning the catalytic triad of 1BWR (Asp192, Ser47, His195) to the putative catalytic triad of 4Q7Q, the RMS is 2.049. (Figure 4) | 1BWR has one chain. When aligning the catalytic triad of 1BWR (Asp192, Ser47, His195) to the putative catalytic triad of 4Q7Q, the RMS is 2.049. (Figure 4) | ||
| - | Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the | + | Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the lower RMS value measured. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. |
== Bacterial Transformation and Plasmid Purification == | == Bacterial Transformation and Plasmid Purification == | ||
| Line 60: | Line 60: | ||
4Q7Q was then purified using the HisPUR Ni-NTA Purification kit. A nickel column as well as equilibration, wash, and elution buffers were used. We then tested for expression using SDS-PAGE, an electrophoresis method that separates proteins by mass in a polyacrylamide gel. BioRad's mini protean tetra protocol was utilized for SDS-PAGE. | 4Q7Q was then purified using the HisPUR Ni-NTA Purification kit. A nickel column as well as equilibration, wash, and elution buffers were used. We then tested for expression using SDS-PAGE, an electrophoresis method that separates proteins by mass in a polyacrylamide gel. BioRad's mini protean tetra protocol was utilized for SDS-PAGE. | ||
| - | Lanes in our gel contain samples of the cell extract, flow through, the third step of washing our protein through the column, as well as all three elutions. We are most interested in seeing our protein eluted in the three elutions. There are bands in these three lanes corresponding to our protein's weight: 87.1 kDa, confirming that we have successfully expressed 4Q7Q. | + | Lanes in our gel contain samples of the cell extract, flow through, the third step of washing our protein through the column, as well as all three elutions. We are most interested in seeing our protein eluted in the three elutions, as the wash steps remove unwanted nucleic acids. There are bands in these three lanes corresponding to our protein's weight: 87.1 kDa, confirming that we have successfully expressed 4Q7Q. |
== pNPB Lipase Assay == | == pNPB Lipase Assay == | ||
Revision as of 02:50, 23 May 2018
4Q7Q
| |||||||||||