We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Jennifer Taylor/Sandbox 4
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
==Characteristics of 4Q7Q== | ==Characteristics of 4Q7Q== | ||
| + | |||
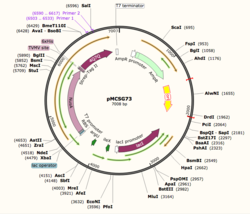
[[Image:plasmid_Map.png|thumb|left|250px|Figure 1: Plasmid Map of pMCSG73, the expression vector for 4Q7Q.]] | [[Image:plasmid_Map.png|thumb|left|250px|Figure 1: Plasmid Map of pMCSG73, the expression vector for 4Q7Q.]] | ||
| Line 19: | Line 20: | ||
== ''In silico'' Analysis == | == ''In silico'' Analysis == | ||
| + | |||
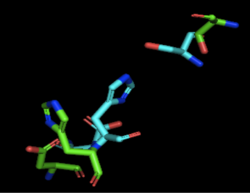
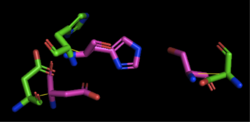
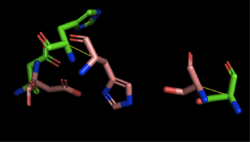
[[Image:4Q7Q_3LIP_alignment.png|thumb|right|250px|Figure 2: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 3LIP's catalytic triad (shown in blue). The RMS is 2.257.]][[Image:4Q7Q_1TAH_alignment.png|thumb|right|250px|Figure 3: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1TAH's catalytic triad (shown in pink). The RMS is 2.205.]][[Image:4Q7Q_1BWR_alignment.png|thumb|right|250px|Figure 4: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1BWR's catalytic triad (shown in pink). The RMS is 2.049.]] | [[Image:4Q7Q_3LIP_alignment.png|thumb|right|250px|Figure 2: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 3LIP's catalytic triad (shown in blue). The RMS is 2.257.]][[Image:4Q7Q_1TAH_alignment.png|thumb|right|250px|Figure 3: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1TAH's catalytic triad (shown in pink). The RMS is 2.205.]][[Image:4Q7Q_1BWR_alignment.png|thumb|right|250px|Figure 4: Alignment of 4Q7Q's putative catalytic triad (shown in green) and 1BWR's catalytic triad (shown in pink). The RMS is 2.049.]] | ||
| Line 35: | Line 37: | ||
Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the lower RMS value measured. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. | Compiling all of the data together, we can see that 1BWR's catalytic triad is most structurally similar to the putative catalytic triad of 4Q7Q due to the lower RMS value measured. Therefore, we hypothesized that 4Q7Q is most likely a hydrolase; through experiments, we can investigate further if 4Q7Q is specifically a lipase. | ||
| + | |||
== Bacterial Transformation and Plasmid Purification == | == Bacterial Transformation and Plasmid Purification == | ||
| - | [[Image:4Q7Q_transformation.png|thumb|left|250px|Figure 5: 4Q7Q bacterial transformation results.]] | ||
| + | [[Image:4Q7Q_transformation.png|thumb|left|250px|Figure 5: 4Q7Q bacterial transformation results.]] | ||
Before characterizing the function of 4Q7Q, we first needed to synthesize the protein through first transcribing 4Q7Q's DNA to amplify it and then translating it to express it. First, 4Q7Q's DNA was transcribed using its expression vector, the plasmid pMCS573. Since transformation must occur within a cell, the plasmid was transformed into DH5α cells using New England Biolabs protocol. | Before characterizing the function of 4Q7Q, we first needed to synthesize the protein through first transcribing 4Q7Q's DNA to amplify it and then translating it to express it. First, 4Q7Q's DNA was transcribed using its expression vector, the plasmid pMCS573. Since transformation must occur within a cell, the plasmid was transformed into DH5α cells using New England Biolabs protocol. | ||
| Line 46: | Line 49: | ||
However, although DH5α cells maximize the efficiency of transformations, they do not contain T7 polymerase, which is essential for protein expression. Therefore, the purified plasmid underwent another bacterial transformation into BL21 (DE3) cells that do contain T7 polymerase using New England Biolabs protocol. | However, although DH5α cells maximize the efficiency of transformations, they do not contain T7 polymerase, which is essential for protein expression. Therefore, the purified plasmid underwent another bacterial transformation into BL21 (DE3) cells that do contain T7 polymerase using New England Biolabs protocol. | ||
| + | |||
== Protein Expression == | == Protein Expression == | ||
| Line 57: | Line 61: | ||
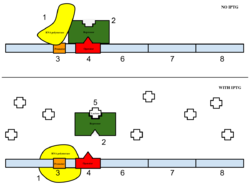
The sample was then induced with IPTG. Essentially, IPTG is a reagent that prevents the repressor from binding to the operator to allow expression to occur (Figure 5). | The sample was then induced with IPTG. Essentially, IPTG is a reagent that prevents the repressor from binding to the operator to allow expression to occur (Figure 5). | ||
| + | |||
== Protein Purification == | == Protein Purification == | ||
| + | |||
[[Image:4Q7Q_gel.png|thumb|left|250px|Figure 7: SDS-PAGE gel. Bands are present at 87.1 kDa.]] | [[Image:4Q7Q_gel.png|thumb|left|250px|Figure 7: SDS-PAGE gel. Bands are present at 87.1 kDa.]] | ||
Revision as of 03:06, 23 May 2018
4Q7Q
| |||||||||||