User:Kayque Alves Telles Silva/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Hfq== | ==Hfq== | ||
<StructureSection load='1kq1' size='400' side='right' scene='78/789833/Hfq/2' caption='Host Factor for the Replication of the Qβ Phage RNA (Hfq)'> | <StructureSection load='1kq1' size='400' side='right' scene='78/789833/Hfq/2' caption='Host Factor for the Replication of the Qβ Phage RNA (Hfq)'> | ||
| - | Hfq is a bacterial cytoplasmatic protein, first named for being a Host Factor for the Replication of the Qβ Phage RNA[1 | + | Hfq is a bacterial cytoplasmatic protein, first named for being a Host Factor for the Replication of the Qβ Phage RNA[1]. Hfq acts pleiotropically and impacts both the degradation and the translation efficiency of mRNAs [2], through the modification of sRNA[3]. Besides, Hfq has a strongly conserved structure[] and belongs to the LSm protein family, present in each of the three domains of life. Despite Hfq’s underscored importance, there is a wide spectrum of cellular functions, as growth rate and cell length, that are directly dependent of its chaperone function and have been discovered in the last century[5,6,7,8], but many others remain unknown. |
| - | == | + | == Function == |
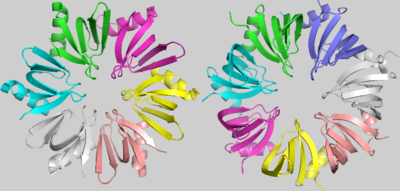
| - | <scene name='78/789833/Hfq/2'>Hfq</scene> is a doughnut shaped homohexamer of roughly 8,9kDa, ~65Å diameter and ~23Å width, as described in S.aureus[1]. The action mechanisms of Hfq as a chaperone are based in its interaction with sRNAs. The best literature described functions among them are: the sequestering of ribosome binding sites (RBS); the inhibition of RBS-blocking mRNA regions, allowing translation; the stabilization of sRNAs which would otherwise be degraded; and the degradation of mRNAs. Hfq might also contribute to the degradation of certain mRNAs directly, but rarely | + | <scene name='78/789833/Hfq/2'>Hfq</scene> is a doughnut shaped homohexamer of roughly 8,9kDa, ~65Å diameter and ~23Å width, as described in S.aureus[1]. The action mechanisms of Hfq as a chaperone are based in its interaction with sRNAs. The best literature described functions among them are: the sequestering of ribosome binding sites (RBS); the inhibition of RBS-blocking mRNA regions, allowing translation; the stabilization of sRNAs which would otherwise be degraded; and the degradation of mRNAs. Hfq might also contribute to the degradation of certain mRNAs directly, but rarely[9]. |
| + | == Structure == | ||
The <scene name='78/789833/Subunit/1'>basic subunit</scene> which forms the homo-hexamer of Hfq has 77 residues[1] comprising one N-terminal α helix (α1), five beta sheets (β1-β5) and one variable region. An unstructured carboxy-terminal region, which may have >100 residues in certain species is also present[10 -Updegrove]. The fold of the beta sheets is characterized by two important <scene name='78/789833/Motifs/5'>motifs</scene>, Sm1 and Sm2, used to clusterize the LSm protein family components considering their highlighted conservation throughout Life’s domains. | The <scene name='78/789833/Subunit/1'>basic subunit</scene> which forms the homo-hexamer of Hfq has 77 residues[1] comprising one N-terminal α helix (α1), five beta sheets (β1-β5) and one variable region. An unstructured carboxy-terminal region, which may have >100 residues in certain species is also present[10 -Updegrove]. The fold of the beta sheets is characterized by two important <scene name='78/789833/Motifs/5'>motifs</scene>, Sm1 and Sm2, used to clusterize the LSm protein family components considering their highlighted conservation throughout Life’s domains. | ||
| Line 16: | Line 17: | ||
The <scene name='78/789833/Subunits_interaction/1'>subunits interaction</scene> is determined by hydrogen bonds between tyrosines present in the β4 and β5 sheets, the Tyr56 and the Tyr63, respectively. The interaction between residues from α1 and the turn between β2 and β3 also contributes to this conformation. The shorter “Variable region” present in Hfq, when compared to the heptameric LSm proteins, allows a closer rotation of the subunits, promoting the hexameric structure of Hfq[1]. | The <scene name='78/789833/Subunits_interaction/1'>subunits interaction</scene> is determined by hydrogen bonds between tyrosines present in the β4 and β5 sheets, the Tyr56 and the Tyr63, respectively. The interaction between residues from α1 and the turn between β2 and β3 also contributes to this conformation. The shorter “Variable region” present in Hfq, when compared to the heptameric LSm proteins, allows a closer rotation of the subunits, promoting the hexameric structure of Hfq[1]. | ||
| + | == RNA Interaction== | ||
The proximal face of Hfq interacts with U-rich 5′-AUUUUUG-3′ RNA sequences, forming a small <scene name='78/789833/Hfq-rna_interaction/2'>Hfq-RNA Interaction</scene> around its pore. Residues such as Tyr42, Lys41 and Gln8 guarantee the bond with uracils and each subunit contacts one nucleotide, except the G residue that remains exposed out of the ring. However, in the predominantly <scene name='78/789833/Rna_distal_face/1'>RNA at distal face</scene>, each subunit bonds with three nucleotides, forming a wider ring of RNA. In this case, A(A/G)N - N being any base - sequences are preferred. | The proximal face of Hfq interacts with U-rich 5′-AUUUUUG-3′ RNA sequences, forming a small <scene name='78/789833/Hfq-rna_interaction/2'>Hfq-RNA Interaction</scene> around its pore. Residues such as Tyr42, Lys41 and Gln8 guarantee the bond with uracils and each subunit contacts one nucleotide, except the G residue that remains exposed out of the ring. However, in the predominantly <scene name='78/789833/Rna_distal_face/1'>RNA at distal face</scene>, each subunit bonds with three nucleotides, forming a wider ring of RNA. In this case, A(A/G)N - N being any base - sequences are preferred. | ||
Revision as of 04:33, 18 June 2018
Hfq
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644