Peptidyl-tRNA hydrolase
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Structural highlights == | == Structural highlights == | ||
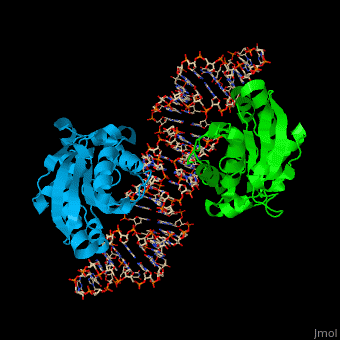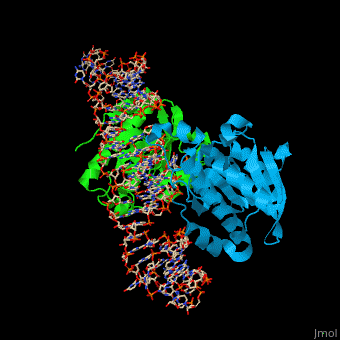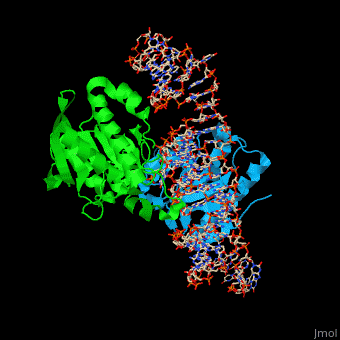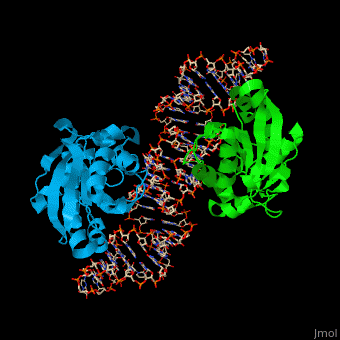
| - | PTH dimer interacts with tRNA at the latter's <scene name='55/551222/Cv/ | + | PTH dimer interacts with tRNA at the latter's <scene name='55/551222/Cv/5'>CCA - the 3' end of the molecule which binds the amino acid</scene>; the <scene name='55/551222/Cv/6'>acceptor site which pass-pairs tRNA N-terminal nucleotides to the C-terminal nucleotides</scene> and the TΨC site (Ψ is pseudouridine) which is the <scene name='55/551222/Cv/7'>tRNA ribosome recognition site</scene><ref>PMID:22923517</ref>. |
</StructureSection> | </StructureSection> | ||
Revision as of 12:19, 31 July 2019
| |||||||||||
3D Structures of peptidyl-tRNA hydrolase
Updated on 31-July-2019
References
- ↑ Das G, Varshney U. Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology. 2006 Aug;152(Pt 8):2191-5. PMID:16849786 doi:http://dx.doi.org/10.1099/mic.0.29024-0
- ↑ Ito K, Murakami R, Mochizuki M, Qi H, Shimizu Y, Miura KI, Ueda T, Uchiumi T. Structural basis for the substrate recognition and catalysis of peptidyl-tRNA hydrolase. Nucleic Acids Res. 2012 Aug 25. PMID:22923517 doi:10.1093/nar/gks790