Pyranose oxidase
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='' size='350' side='right' caption='Pyranose oxidase complex with FAD and deoxy-fluoro-glucopyranose (PDB code [[3k4l]])' scene='70/708805/Cv/1'> | <StructureSection load='' size='350' side='right' caption='Pyranose oxidase complex with FAD and deoxy-fluoro-glucopyranose (PDB code [[3k4l]])' scene='70/708805/Cv/1'> | ||
== Function == | == Function == | ||
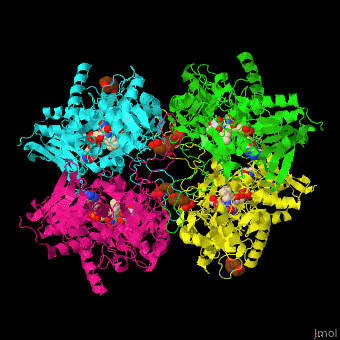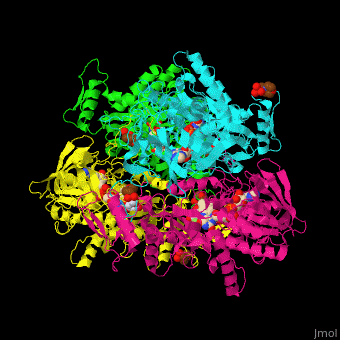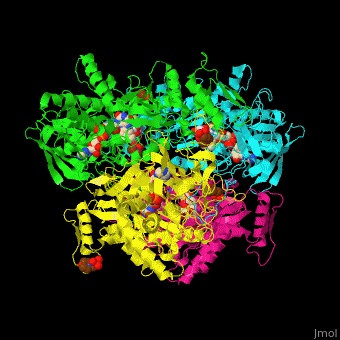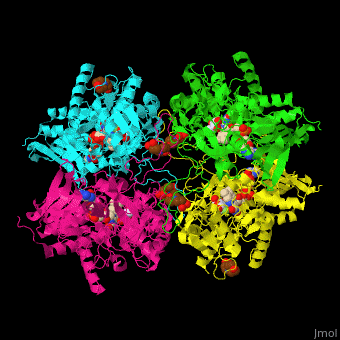
| - | '''Pyranose oxidase''' or '''pyranose 2-oxidase''' (P2O) catalyzes the conversion of D-glucose and molecular oxygen to 2-dehydro-D-glucose and H<sub>2</sub>O<sub>2</sub>. It is abundant in lignin-degrading white rot fungi. P2O is a <scene name='70/708805/Cv/ | + | '''Pyranose oxidase''' or '''pyranose 2-oxidase''' (P2O) catalyzes the conversion of D-glucose and molecular oxygen to 2-dehydro-D-glucose and H<sub>2</sub>O<sub>2</sub>. It is abundant in lignin-degrading white rot fungi. P2O is a <scene name='70/708805/Cv/7'>tetramer</scene> flavoprotein containing the <scene name='70/708805/Cv/8'>prosthetic group FAD in each monomer</scene>. P2O oxidizes several aldopyranoses with the preferred electron donors being D-glucose, D-xylose and D-sorbose<ref>PMID:11152063</ref>. |
== Relevance == | == Relevance == | ||
| Line 8: | Line 8: | ||
== Structural highlights == | == Structural highlights == | ||
| - | The <scene name='70/708805/Cv/ | + | The <scene name='70/708805/Cv/9'>active site of P2O</scene> is gated by a highly conserved loop which determines the substrate specificity. Water molecules are shown as red spheres. The active site contains the FAD cofactor<ref>PMID:20528921</ref>. |
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||
3D Structures of pyranose oxidase
Updated on 21-August-2019
References
- ↑ Giffhorn F. Fungal pyranose oxidases: occurrence, properties and biotechnical applications in carbohydrate chemistry. Appl Microbiol Biotechnol. 2000 Dec;54(6):727-40. PMID:11152063
- ↑ Spadiut O, Tan TC, Pisanelli I, Haltrich D, Divne C. Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase. FEBS J. 2010 Jul;277(13):2892-909. Epub 2010 Jun 2. PMID:20528921 doi:10.1111/j.1742-4658.2010.07705.x