We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Shikimate dehydrogenase
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
==Structural highlights == | ==Structural highlights == | ||
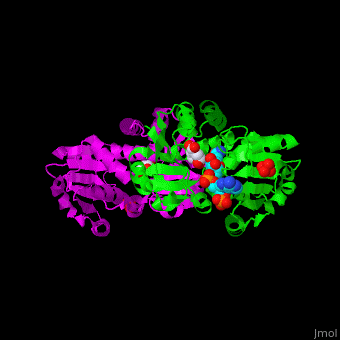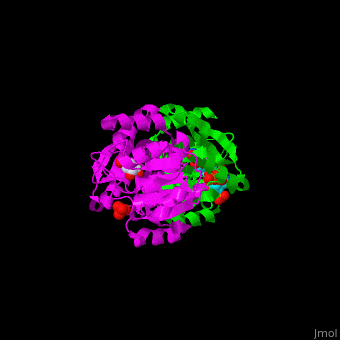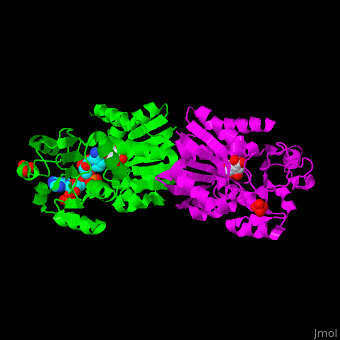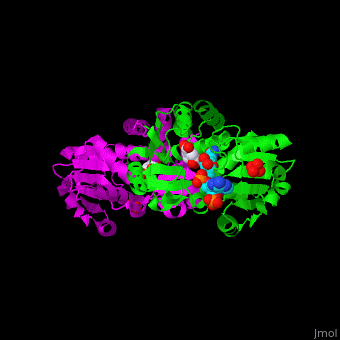
AroE changes its conformation from an open to closed one upon binding of the cofactor NADP. The active site contains the substrate shikimate<ref>PMID:17825835</ref>. | AroE changes its conformation from an open to closed one upon binding of the cofactor NADP. The active site contains the substrate shikimate<ref>PMID:17825835</ref>. | ||
| - | *<scene name='54/540162/Cv/ | + | *<scene name='54/540162/Cv/5'>NADP binding site</scene>. Water molecules are shown as red spheres. |
| - | *<scene name='54/540162/Cv/ | + | *<scene name='54/540162/Cv/6'>Whole binding site</scene>. |
</StructureSection> | </StructureSection> | ||
== 3D Structures of shikimate dehydrogenase == | == 3D Structures of shikimate dehydrogenase == | ||
Revision as of 13:14, 11 September 2019
| |||||||||||
3D Structures of shikimate dehydrogenase
Updated on 11-September-2019
References
- ↑ Singh S, Stavrinides J, Christendat D, Guttman DS. A phylogenomic analysis of the shikimate dehydrogenases reveals broadscale functional diversification and identifies one functionally distinct subclass. Mol Biol Evol. 2008 Oct;25(10):2221-32. doi: 10.1093/molbev/msn170. Epub 2008 Jul, 31. PMID:18669580 doi:http://dx.doi.org/10.1093/molbev/msn170
- ↑ Bagautdinov B, Kunishima N. Crystal structures of shikimate dehydrogenase AroE from Thermus thermophilus HB8 and its cofactor and substrate complexes: insights into the enzymatic mechanism. J Mol Biol. 2007 Oct 19;373(2):424-38. Epub 2007 Aug 21. PMID:17825835 doi:10.1016/j.jmb.2007.08.017
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Sohail Jooma