This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Rde 4 sandbox
From Proteopedia
| Line 9: | Line 9: | ||
== Structure of RDE-4 == | == Structure of RDE-4 == | ||
| - | RDE-4 has five major regions: an <scene name='79/798389/N-terminal_of_rde-4/1'>N-terminal region (residues 1-43)</scene>, two dsRBDs <scene name='79/798389/Dsrbd_residues_44-108/1'>(residues 44-108)</scene> and <scene name='79/798389/Dsrbd_residues_170-235/2'>(residues 170-235)</scene>, a <scene name='79/798389/Long_linker_of_rde-4/1'>Long Linker (residues 109-169)</scene>, and a C-terminal domain (residues 236–385). Amino acid residues 1–32 and 136–151 adopt a random coiled structure and do not make any contact within itself or with the rest of the structure. Parts of the linker, residues 109–135 and 152–169, assume an α-helix and an extended loop structure, respectively. There is not any long-range nuclear Overhauser effect information between regions 1–135 and 152– 243, suggesting that the unstructured part of the linker 136–151 separates both of these regions. The region encompassed by 236–243 folds into an α-helix. The amino acid regions 44–108 and 170–235 form dsRBD1 and dsRBD2, respectively. The hydrophobic residues Leu45, Val47, Leu48, Val55, Trp62, Met73, Leu75, Leu77, Ile80, Val82, Leu101, and Val105 stabilize dsRBD1, and Val171, Leu174, Leu183, Val201, Met205, Met227, and Leu232 form the core of dsRBD2. Apart from the canonical dsRBD fold, there are additional secondary-structural elements that are observed in both RDE-4D1 and RDE-4D2. <ref name=second> | + | RDE-4 has five major regions: an <scene name='79/798389/N-terminal_of_rde-4/1'>N-terminal region (residues 1-43)</scene>, two dsRBDs <scene name='79/798389/Dsrbd_residues_44-108/1'>(residues 44-108)</scene> and <scene name='79/798389/Dsrbd_residues_170-235/2'>(residues 170-235)</scene>, a <scene name='79/798389/Long_linker_of_rde-4/1'>Long Linker (residues 109-169)</scene>, and a C-terminal domain (residues 236–385). Amino acid residues 1–32 and 136–151 adopt a random coiled structure and do not make any contact within itself or with the rest of the structure. Parts of the linker, residues 109–135 and 152–169, assume an α-helix and an extended loop structure, respectively. There is not any long-range nuclear Overhauser effect information between regions 1–135 and 152– 243, suggesting that the unstructured part of the linker 136–151 separates both of these regions. The region encompassed by 236–243 folds into an α-helix. The amino acid regions 44–108 and 170–235 form dsRBD1 and dsRBD2, respectively. The hydrophobic residues Leu45, Val47, Leu48, Val55, Trp62, Met73, Leu75, Leu77, Ile80, Val82, Leu101, and Val105 stabilize dsRBD1, and Val171, Leu174, Leu183, Val201, Met205, Met227, and Leu232 form the core of dsRBD2. Apart from the canonical dsRBD fold, there are additional secondary-structural elements that are observed in both RDE-4D1 and RDE-4D2. <ref name=second>DOI: 10.1042/BJ20131347</ref> |
== Function == | == Function == | ||
In C. elegans, exogenous dsRNA is detected and bound by RDE-4, which stimulates dicer activity. RDE-4 initiates the siRNA pathway by binding to long dsRNA and assisting Dcr-1, a Dicer1 homologue, to facilitate siRNA production. RDE-4 interacts with Dcr-1, RDE-1, DRH-1 (Dicer-related helicase 1) and long dsRNA, which thereby suggests that RDE-4 is required only for the initiation of the RNAi pathway to generate siRNA, although high levels of dsRNA abrogate RDE-4’s role in the RNAi initiation. RDE-4 co- operatively binds to long dsRNA with nanomolar affinity, but exhibits micromolar affinity for short dsRNA, and the enhanced affinity arises from binding of several RDE-4 molecules to a single long dsRNA. <ref name=third>DOI: 10.1016/j.jmb.2008.10.002</ref> The C-terminal region of RDE-4 is necessary and sufficient to induce homodimerization RDE-4 linker– dsRBD2 are absolutely essential in C. elegans gene silencing and RDE-4 dsRBD1 and the C-terminal domain do not have any primary role in vivo. RDE-4 recognizes the A-form of nucleic acid structure adopted by dsRNA and the interaction is sequence-independent.<ref name=second/> RDE-4 dimerization is important for the assembly of active RDE-4/Dicer complexes via one of two proposed scenarios, proper Dicer recruitment to dsRNA, or facilitating Dicer dimerization. <ref name=first/> | In C. elegans, exogenous dsRNA is detected and bound by RDE-4, which stimulates dicer activity. RDE-4 initiates the siRNA pathway by binding to long dsRNA and assisting Dcr-1, a Dicer1 homologue, to facilitate siRNA production. RDE-4 interacts with Dcr-1, RDE-1, DRH-1 (Dicer-related helicase 1) and long dsRNA, which thereby suggests that RDE-4 is required only for the initiation of the RNAi pathway to generate siRNA, although high levels of dsRNA abrogate RDE-4’s role in the RNAi initiation. RDE-4 co- operatively binds to long dsRNA with nanomolar affinity, but exhibits micromolar affinity for short dsRNA, and the enhanced affinity arises from binding of several RDE-4 molecules to a single long dsRNA. <ref name=third>DOI: 10.1016/j.jmb.2008.10.002</ref> The C-terminal region of RDE-4 is necessary and sufficient to induce homodimerization RDE-4 linker– dsRBD2 are absolutely essential in C. elegans gene silencing and RDE-4 dsRBD1 and the C-terminal domain do not have any primary role in vivo. RDE-4 recognizes the A-form of nucleic acid structure adopted by dsRNA and the interaction is sequence-independent.<ref name=second/> RDE-4 dimerization is important for the assembly of active RDE-4/Dicer complexes via one of two proposed scenarios, proper Dicer recruitment to dsRNA, or facilitating Dicer dimerization. <ref name=first/> | ||
| + | |||
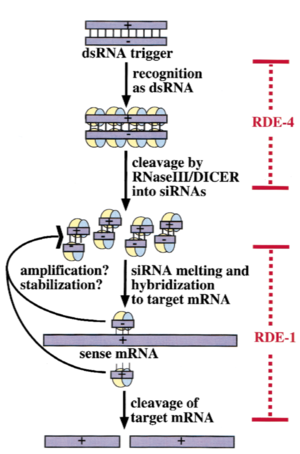
| + | [[Image:Image-RDE-4 Mechanism.png|300px|left|thumb| A working model showing roles of RDE-1, RDE-4, and siRNAs in the interference reaction]] <ref name=fourth>DOI: 10.1017.S1355838201011074</ref> | ||
| + | |||
== Importance == | == Importance == | ||
Revision as of 03:28, 9 October 2018
Contents |
Introduction
RDE-4 was identified through a genome-wide screening as a gene responsible for greatly reducing or abolishing RNAi in C. elegans. In organisms ranging from Arabidopsis to humans, Dicer requires dsRNA-binding proteins (dsRBPs) to carry out its roles in RNA interference (RNAi) and micro-RNA (miRNA) processing. In Caenorhabditis elegans, the dsRBP RDE-4 acts with Dicer during the initiation of RNAi, when long dsRNA is cleaved to small interfering RNAs (siRNAs). RDE-4 is not sequence-specific, however, RDE-4 binds with higher affinity to long dsRNA. Interestingly, RDE- 4 is the only protein that exhibits micromolar affinity for dsRNA in the absence of co-operativity. In addition, RDE-4 is a homodimer in solution. [1]
|
|
Structure of RDE-4
RDE-4 has five major regions: an , two dsRBDs and , a , and a C-terminal domain (residues 236–385). Amino acid residues 1–32 and 136–151 adopt a random coiled structure and do not make any contact within itself or with the rest of the structure. Parts of the linker, residues 109–135 and 152–169, assume an α-helix and an extended loop structure, respectively. There is not any long-range nuclear Overhauser effect information between regions 1–135 and 152– 243, suggesting that the unstructured part of the linker 136–151 separates both of these regions. The region encompassed by 236–243 folds into an α-helix. The amino acid regions 44–108 and 170–235 form dsRBD1 and dsRBD2, respectively. The hydrophobic residues Leu45, Val47, Leu48, Val55, Trp62, Met73, Leu75, Leu77, Ile80, Val82, Leu101, and Val105 stabilize dsRBD1, and Val171, Leu174, Leu183, Val201, Met205, Met227, and Leu232 form the core of dsRBD2. Apart from the canonical dsRBD fold, there are additional secondary-structural elements that are observed in both RDE-4D1 and RDE-4D2. [2]
Function
In C. elegans, exogenous dsRNA is detected and bound by RDE-4, which stimulates dicer activity. RDE-4 initiates the siRNA pathway by binding to long dsRNA and assisting Dcr-1, a Dicer1 homologue, to facilitate siRNA production. RDE-4 interacts with Dcr-1, RDE-1, DRH-1 (Dicer-related helicase 1) and long dsRNA, which thereby suggests that RDE-4 is required only for the initiation of the RNAi pathway to generate siRNA, although high levels of dsRNA abrogate RDE-4’s role in the RNAi initiation. RDE-4 co- operatively binds to long dsRNA with nanomolar affinity, but exhibits micromolar affinity for short dsRNA, and the enhanced affinity arises from binding of several RDE-4 molecules to a single long dsRNA. [3] The C-terminal region of RDE-4 is necessary and sufficient to induce homodimerization RDE-4 linker– dsRBD2 are absolutely essential in C. elegans gene silencing and RDE-4 dsRBD1 and the C-terminal domain do not have any primary role in vivo. RDE-4 recognizes the A-form of nucleic acid structure adopted by dsRNA and the interaction is sequence-independent.[2] RDE-4 dimerization is important for the assembly of active RDE-4/Dicer complexes via one of two proposed scenarios, proper Dicer recruitment to dsRNA, or facilitating Dicer dimerization. [1]
[4]
Importance
The RDE-4 protein, and its interactions with RDE-1 and Dicer, is rather important due to the fact that it cleaves the dsRNA to form small interfering RNA or
References
- ↑ 1.0 1.1 Parker GS, Eckert DM, Bass BL. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006 May;12(5):807-18. doi: 10.1261/rna.2338706. Epub 2006 Apr 7. PMID:16603715 doi:http://dx.doi.org/10.1261/rna.2338706
- ↑ 2.0 2.1 Chiliveri SC, Deshmukh MV. Structure of RDE-4 dsRBDs and mutational studies provide insights in the dsRNA recognition in C. elegans RNAi pathway. Biochem J. 2013 Nov 21. PMID:24256178 doi:http://dx.doi.org/10.1042/BJ20131347
- ↑ Parker GS, Maity TS, Bass BL. dsRNA binding properties of RDE-4 and TRBP reflect their distinct roles in RNAi. J Mol Biol. 2008 Dec 26;384(4):967-79. doi: 10.1016/j.jmb.2008.10.002. Epub 2008 , Oct 11. PMID:18948111 doi:http://dx.doi.org/10.1016/j.jmb.2008.10.002
- ↑ doi: https://dx.doi.org/10.1017.S1355838201011074