User:Gisele A. Andree/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
==Structure of Eukaryotic Dihydropyrimidine Dehydrogenase == | ==Structure of Eukaryotic Dihydropyrimidine Dehydrogenase == | ||
<StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | <StructureSection load='1stp' size='340' side='right' caption='Caption for this structure' scene=''> | ||
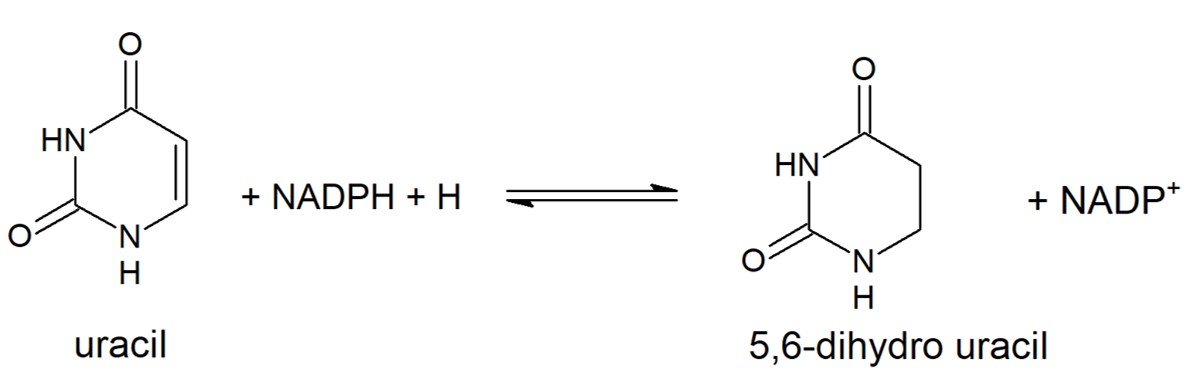
| - | + | Dihydropyrimidine Dehydrogenase (DPD) is an important enzyme involved in the degradation of pyrimidines in the body. It is a 220 kDa protein that binds co-factors; FAD, FMN, [4Fe-4S] clusters, and substrates; NADPH and pyrimidines. | |
| + | |||
You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| Line 11: | Line 12: | ||
== Disease == | == Disease == | ||
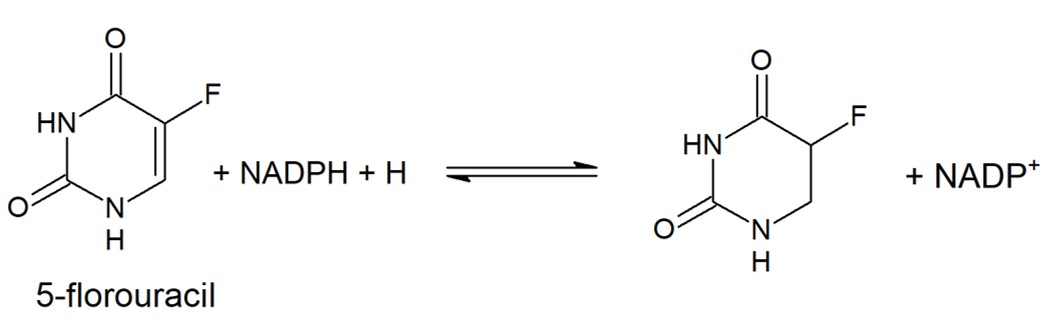
| - | 5-fluorouracil (5-FU) is a drug used to treat a variety of cancers as it has wide anti-tumor activity & works well alongside other chemotherapy drugs. In the human liver 80-85% of 5-FU is catabolized into inactive, and potentially toxic, metabolites by DPD | + | 5-fluorouracil (5-FU) is a drug used to treat a variety of cancers as it has wide anti-tumor activity & works well alongside other chemotherapy drugs. In the human liver 80-85% of 5-FU is catabolized into inactive, and potentially toxic, metabolites by DPD. |
| - | + | [[Image:5FU_reduction_.jpg]] | |
| + | Only 1-3% of the original dose proceeds through anabolic pathways to create active cytotoxic complexes. The active complexes inhibit DNA synthesis and the processing and function of RNA processing thus producing a deleterious effect on both healthy and cancerous cells. | ||
| + | |||
| + | DPD decreases effectivity of drug thus requires a very high dosages, leading to major side effects. Luckily, inhibitors are in development and some are in clinical trials. | ||
| - | == Relevance == | ||
== Structural highlights == | == Structural highlights == | ||
| + | DPD is a homodimer, with each monomer consisting of five domains. | ||
| + | |||
| + | '''Domain 1''' | ||
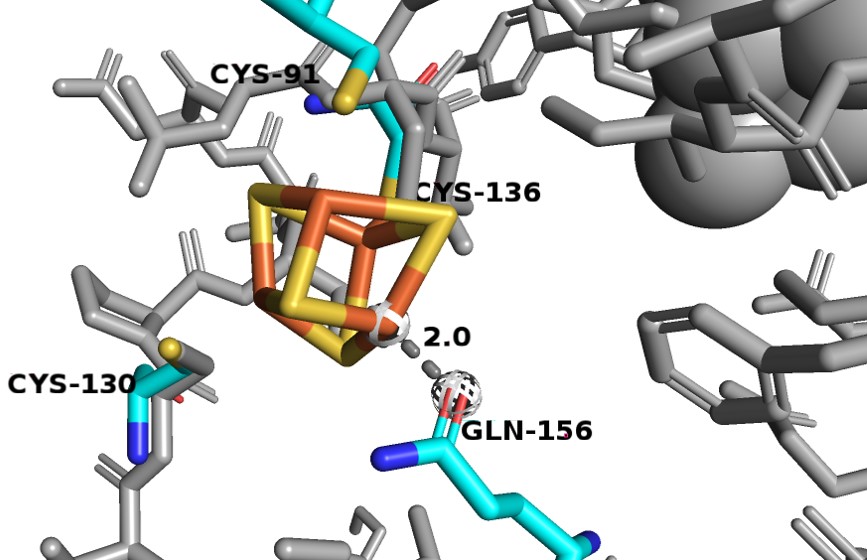
| + | Consists of residues 27-173. It binds 2 Fe-S clusters and is made of exclusively alpha-helices. One of the [4Fe-4S] clusters is odd as it has a different coordination that is not observed in other Fe-S proteins. Three Fe atoms interact with cysteines 91, 130 and 136, but the fourth Fe atom interacts with the Oε1 glutamine 156. Since the oxygen has a smaller atomic radius than sulfur, so the Fe-O distance is much shorter (~2.0 Å) than the average Fe-S interactions seen in DPD (2.3 Å). | ||
| + | |||
| + | [[Image:D1_Fe.jpg]] | ||
| + | |||
| + | '''Domain 2 and 3''' | ||
| + | Domain 2 consists of residues 173-286 and 442-524. It binds FAD and NADPH. It contains a central parallel beta-sheet surrounded by alpha helices, forming Rossman-type nucleotide binding motifs that bind FAD and NADPH. | ||
| + | |||
| + | Domain 3 consists of residues 287-441 and also binds FAD and NADPH. Just like domain 2, contains a central parallel beta-sheet surrounded by alpha helices, forming Rossman-type nucleotide binding motifs that bind FAD and NADPH. But, D3 also contains an additional 3 stranded antiparallel beta-sheet. With the aforementioned exception, D2 and D3 are so similar that they are thought to have originated from gene duplication. | ||
| + | |||
| + | In the following figure, domain 2 is shown in blue, domain 3 is in green, FAD is colored in orange and NADPH is in pink. | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
| - | < | + | <Dobritzsch, D., Schneider, G., Schnackerz, K. D., & Lindqvist, Y. (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti‐cancer drug 5‐fluorouracil. The EMBO journal, 20(4), 650-660./> |
| + | <Schnackerz, K. D., Dobritzsch, D., Lindqvist, Y., & Cook, P. F. (2004). Dihydropyrimidine dehydrogenase: a flavoprotein with four iron–sulfur clusters. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1701(1-2), 61-74./> | ||
Revision as of 20:00, 13 December 2018
Structure of Eukaryotic Dihydropyrimidine Dehydrogenase
| |||||||||||
References
<Dobritzsch, D., Schneider, G., Schnackerz, K. D., & Lindqvist, Y. (2001). Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti‐cancer drug 5‐fluorouracil. The EMBO journal, 20(4), 650-660./> <Schnackerz, K. D., Dobritzsch, D., Lindqvist, Y., & Cook, P. F. (2004). Dihydropyrimidine dehydrogenase: a flavoprotein with four iron–sulfur clusters. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1701(1-2), 61-74./>