We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1489
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
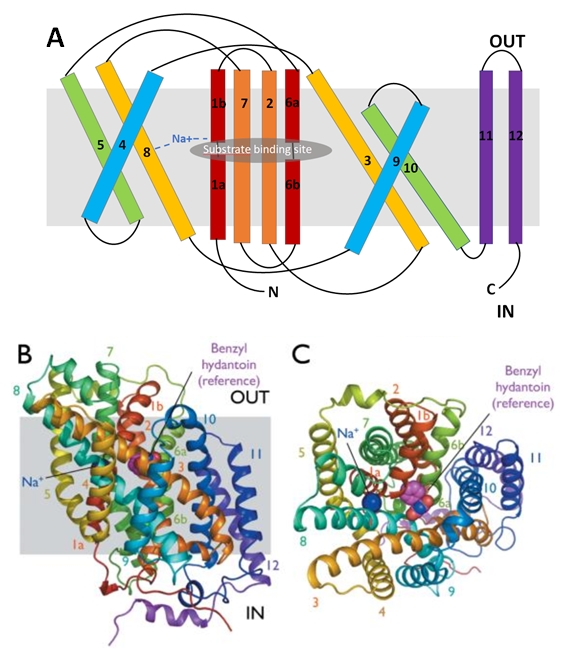
The substrate- and cation-binding sites are located in the space between the central four-helix-bundle and the outer helix layer. | The substrate- and cation-binding sites are located in the space between the central four-helix-bundle and the outer helix layer. | ||
| - | == Structure of the substrate | + | == Structure of the substrate binding site == |
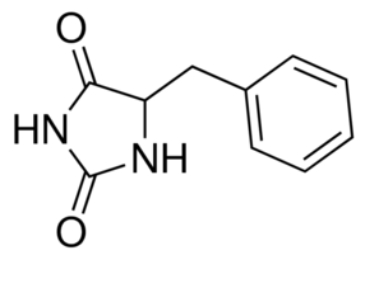
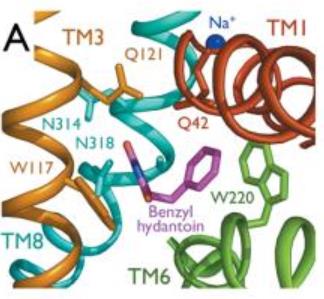
The substrate binding site is located at the break of the TMs 1 and 6. | The substrate binding site is located at the break of the TMs 1 and 6. | ||
| Line 35: | Line 35: | ||
Figure 3 : Substrate binding site | Figure 3 : Substrate binding site | ||
| + | |||
| + | == Structure of the cation binding site == | ||
| + | |||
| + | Mhp1 is a sodium dependent protein. The sodium binds at the C-terminal end of TM1a and interacts with TM8 (Figure 2.A). The dipole moment at the C-terminus of TM1a contributes to the binding. | ||
| + | Experiments have shown that benzyl-hydantoin increases the affinity of sodium for Mhp1 and reciprocally sodium increases the affinity of benzyl-hydantoin for Mhp1. Therefore, the binding of the substrate and the cation are closely coupled. | ||
| + | |||
| + | |||
| + | == Conformational states == | ||
| + | |||
| + | Mhp1 exists in two conformational states depending if the substrate is bound or not: the substrate free structure (-BH), corresponding to the outward-facing open and the substrate bound structure (+BH), corresponding to the outward-facing occluded (Figure 4). | ||
| + | |||
| + | [[Image:Example.jpg]] | ||
| + | |||
| + | Figure 4: The conformational change upon the substrate binding | ||
| + | |||
| + | The binding of the substrate induces conformational changes of the protein allowing it uptake in the cell. | ||
| + | |||
| + | [[Image:Example.jpg]] | ||
| + | |||
| + | Figure 5 : Proposed substrate translocation mechanism by Mhp1 | ||
| + | |||
| + | |||
| + | The binding of the substrate in the binding site leads to a switch from the outward-facing open state to the outward-facing occluded state. The TM10 arrangement changes (Figure 4.C) and closes the access to the “OUT” side space of the membrane (Figure 5.A). | ||
| + | |||
| + | Then, there is a change from the outward-facing occluded state to the inward-facing occluded state. (Figure 5.B) The substrate-binding site is occluded from the inside of the membrane. It seems that the movement involves the helix bundle of TMs 3 and 8. Moreover, researchers are working on the possibility of a coordinated shifting of TMs 1 and 6 shift with TMs 3 and 8. | ||
| + | Eventually, there is a switch from the inward-facing occluded state to the inward-facing open state. This allows the release of the substrate in the cytoplasm. However, the structures involved in the change still be unclear (Figure 5.C). | ||
| + | |||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 11:31, 9 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2JLN
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644