Sandbox Reserved 1491
From Proteopedia
(Difference between revisions)
| Line 42: | Line 42: | ||
The sequence of the domain has been particularly preserved around OGA (when the protein is folded)<ref>http://consurf.tau.ac.il/fgij/fg.htm?mol=/temp/2XMLA_ConSurf_DB_pipe.pdb </ref>. Thus, the 3D structure has been very preserved as well, indicating that the structure around OGA is essential. | The sequence of the domain has been particularly preserved around OGA (when the protein is folded)<ref>http://consurf.tau.ac.il/fgij/fg.htm?mol=/temp/2XMLA_ConSurf_DB_pipe.pdb </ref>. Thus, the 3D structure has been very preserved as well, indicating that the structure around OGA is essential. | ||
| - | == | + | == Epigenetics == |
| - | Specific enzymes are directly involved in the modification of genes expression without altering the nucleotide sequence. They can modify the chromatin structure by adding (writers), reading (readers) or removing (erasers) marks : | + | Specific enzymes are directly involved in the '''modification of genes expression''' without altering the nucleotide sequence. They can modify the chromatin structure by '''adding''' (writers), '''reading''' (readers) or '''removing''' (erasers) marks : acetyl, methyl, phosphoryl groups, ubiquitin<ref>Kupershmit, Ilana, Hanan Khoury-Haddad, Samah W. Awwad, Noga Guttmann-Raviv, and Nabieh Ayoub. “KDM4C (GASC1) Lysine Demethylase Is Associated with Mitotic Chromatin and Regulates Chromosome Segregation during Mitosis.” Nucleic Acids Research 42, no. 10 (June 2, 2014): 6168–82. https://doi.org/10.1093/nar/gku253.</ref>... These [https://en.wikipedia.org/wiki/Epigenetics epigenetic]marks can either make the gene sequence more or less accessible depending on their nature, histones and labelled amino acids. All combinations of the nature and the localization of the marks form the histone code. |
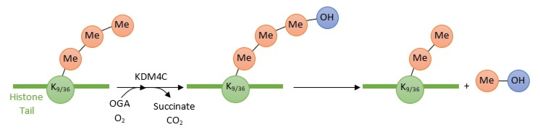
KDM4C is a histone demethylase. This newly discovered class of proteins plays a central role in the histone modifications : it removes the methyl group (which is very stable) from the epigenetically modified amino acid. Its actions has directed consequences on gene expression. By removing repressive histone marks (H3K9me3 and H3K36me3) from target genes KDM4C promotes the formation of euchromatin and therefore transcriptional activation. | KDM4C is a histone demethylase. This newly discovered class of proteins plays a central role in the histone modifications : it removes the methyl group (which is very stable) from the epigenetically modified amino acid. Its actions has directed consequences on gene expression. By removing repressive histone marks (H3K9me3 and H3K36me3) from target genes KDM4C promotes the formation of euchromatin and therefore transcriptional activation. | ||
Revision as of 16:41, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
2xml - KDM4C catalytic domain
2xml is a 2 chain structure. This domain belongs to the Human KDM4C protein.
KDM4C is a histone demethylase involved in the specific demethylation of trimethylated residues (Lys 9 and Lys 36 of histone 3). These marks are specific tags for genes expression modification. KDM4C plays a main role in the modification of cell cycle genes expression and thus involved in the growth of tumoral cells.
| |||||||||||
References
- ↑ Tamaru, H. “Confining Euchromatin/Heterochromatin Territory: Jumonji Crosses the Line.” Genes & Development 24, no. 14 (July 15, 2010): 1465–78. https://doi.org/10.1101/gad.1941010.
- ↑ Nasir Javaid, and Sangdun Choi. “Acetylation- and Methylation-Related Epigenetic Proteins in the Context of Their Targets.” Genes 8, no. 8 (August 7, 2017): 196. https://doi.org/10.3390/genes8080196
- ↑ Shi, Y. G., and Y.-i. Tsukada. “The Discovery of Histone Demethylases.” Cold Spring Harbor Perspectives in Biology 5, no. 9 (September 1, 2013): a017947–a017947. https://doi.org/10.1101/cshperspect.a017947.
- ↑ Labbé, Roselyne M., Andreana Holowatyj, and Zeng-Quan Yang. “Histone Lysine Demethylase (KDM) Subfamily 4: Structures, Functions and Therapeutic Potential.” American Journal of Translational Research 6, no. 1 (2013): 1–15
- ↑ Leurs, Ulrike, Brian Lohse, Kasper D. Rand, Shonoi Ming, Erik S. Riise, Philip A. Cole, Jesper L. Kristensen, and Rasmus P. Clausen. “Substrate- and Cofactor-Independent Inhibition of Histone Demethylase KDM4C.” ACS Chemical Biology 9, no. 9 (September 19, 2014): 2131–38. https://doi.org/10.1021/cb500374f.
- ↑ http://consurf.tau.ac.il/fgij/fg.htm?mol=/temp/2XMLA_ConSurf_DB_pipe.pdb
- ↑ Kupershmit, Ilana, Hanan Khoury-Haddad, Samah W. Awwad, Noga Guttmann-Raviv, and Nabieh Ayoub. “KDM4C (GASC1) Lysine Demethylase Is Associated with Mitotic Chromatin and Regulates Chromosome Segregation during Mitosis.” Nucleic Acids Research 42, no. 10 (June 2, 2014): 6168–82. https://doi.org/10.1093/nar/gku253.
- ↑ Douglas Hanahan et Robert A. Weinberg, « The hallmarks of cancer », Cell, vol. 100, 7 janvier 2000, p. 57-70 (PMID 10647931)
- ↑ https://en.wikipedia.org/wiki/Cancer
- ↑ Gregory, Brittany L., and Vivian G. Cheung. ‘Natural Variation in the Histone Demethylase, KDM4C, Influences Expression Levels of Specific Genes Including Those That Affect Cell Growth’. Genome Research 24, no. 1 (January 2014): 52–63. https://doi.org/10.1101/gr.156141.113
- ↑ Garcia, Jeison, and Fernando Lizcano. ‘KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer’. Breast Cancer : Basic and Clinical Research 10 (2 November 2016): 169–75. https://doi.org/10.4137/BCBCR.S40182.