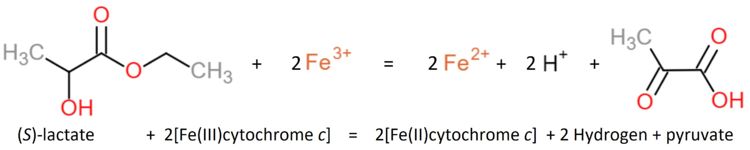Sandbox Reserved 1501
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
The Yeasts L-Lactate Dehydrogenase can be inhibited by heavy metals, oxygen, glycerate, oxalate, malate, phenylpyruvate and fatty acids [Nygaard 1963]<ref>PMID:21638687</ref>. | The Yeasts L-Lactate Dehydrogenase can be inhibited by heavy metals, oxygen, glycerate, oxalate, malate, phenylpyruvate and fatty acids [Nygaard 1963]<ref>PMID:21638687</ref>. | ||
The Enzyme shows a specificity for L-lactate but none for the D-isomer or -hydroxybutyrate. | The Enzyme shows a specificity for L-lactate but none for the D-isomer or -hydroxybutyrate. | ||
| - | + | <!-- hallo --> | |
== Relevance == | == Relevance == | ||
Revision as of 17:50, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ NYGAARD AP. Various forms of D- and L-lactate dehydrogenases in yeast. Ann N Y Acad Sci. 1961 Nov 2;94:774-9. PMID:14480786
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644