We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1502
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
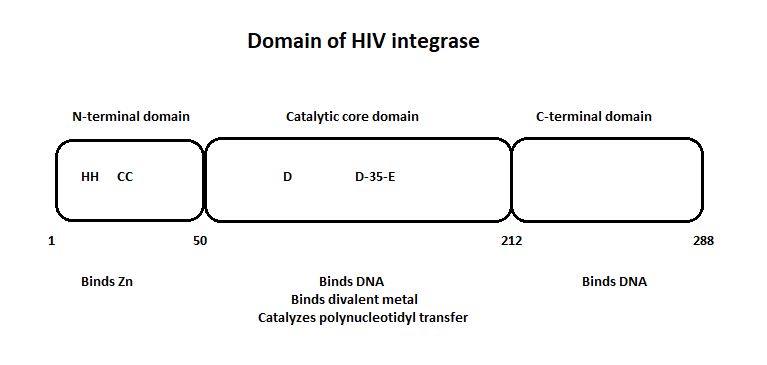
HIV-1 integrase is composed of three domains: the N-terminal (residues 1-49), the core domain (residues 50-212) and the C-terminal domain (residues 213-288). The core domain is responsible for the catalytic activity of the enzyme. It contains three acidic residues, the D,D-35-E motif which plays a key role in catalysis. The N-terminal domain includes the conserved HHCC motif, which binds zinc. The C-terminal domain is less well conserved. [http://www.jbc.org/content/276/26/23213/F2.expansion.html] | HIV-1 integrase is composed of three domains: the N-terminal (residues 1-49), the core domain (residues 50-212) and the C-terminal domain (residues 213-288). The core domain is responsible for the catalytic activity of the enzyme. It contains three acidic residues, the D,D-35-E motif which plays a key role in catalysis. The N-terminal domain includes the conserved HHCC motif, which binds zinc. The C-terminal domain is less well conserved. [http://www.jbc.org/content/276/26/23213/F2.expansion.html] | ||
| - | The complex with the two DNA are displayed on the proteopedia page [[5u1c]]. | ||
====Structure and role of the core domain in the integration==== | ====Structure and role of the core domain in the integration==== | ||
| Line 41: | Line 40: | ||
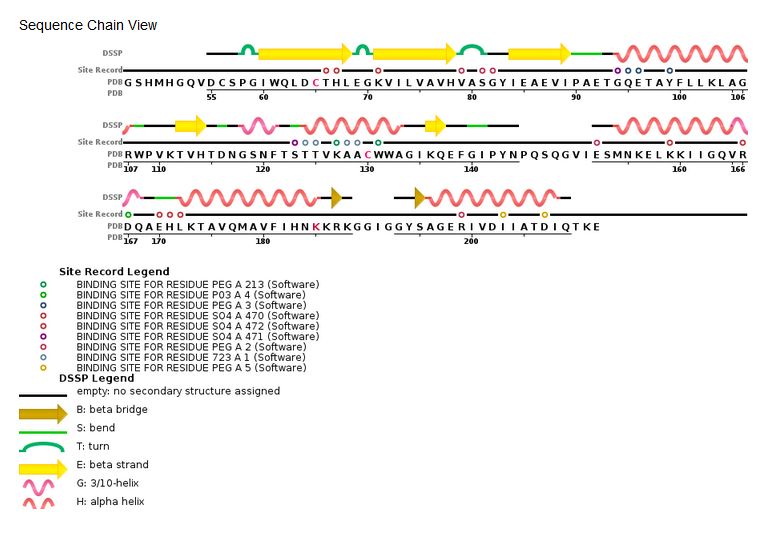
[[Image:HIV Integrase sequence.jpg]] | [[Image:HIV Integrase sequence.jpg]] | ||
| + | |||
| + | ====Structure and role of the core domain==== | ||
| + | |||
| + | |||
| + | ====Structure and role of the N ter domain==== | ||
| + | |||
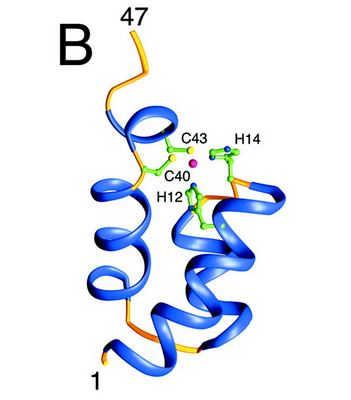
| + | The N-terminal domain of HIV-1 integrase contains a conserved pair of His and Cys residues, a motif similar to the zinc-coordinating residues of zinc fingers. This domain binds zinc but its structure is totally different from that of zinc fingers. It consists of a bundle of three α-helices. It has an SH3 fold, although there is no known functional relationship with the SH3 domains of other proteins. | ||
| + | |||
| + | [[Image:Nter-domain.jpg]] | ||
====Structure and interaction with the antiviral==== | ====Structure and interaction with the antiviral==== | ||
| Line 64: | Line 72: | ||
==See also== | ==See also== | ||
http://proteopedia.org/wiki/index.php/2b4j :integrase ligated with the cofactor LEDGF/p75 | http://proteopedia.org/wiki/index.php/2b4j :integrase ligated with the cofactor LEDGF/p75 | ||
| - | http://proteopedia.org/wiki/index.php/5u1c : integrase complex with the two DNAs | ||
== References == | == References == | ||
Revision as of 22:09, 10 January 2019
| This Sandbox is Reserved from 06/12/2018, through 30/06/2019 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1480 through Sandbox Reserved 1543. |
To get started:
More help: Help:Editing |
3lpt - HIV integrase
| |||||||||||