User:Estelle Blochouse/ Sandbox 1497
From Proteopedia
(Difference between revisions)
| Line 27: | Line 27: | ||
</table> | </table> | ||
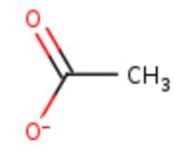
| - | <table><tr><td colspan='2'>In the chain, 5 ligands are present: an acetate ion and 4 copper ions. <br> | + | <table><tr><td colspan='2'>In the chain, 5 ligands are present: an acetate ion (C2H3O2) and 4 copper ions. <br> |
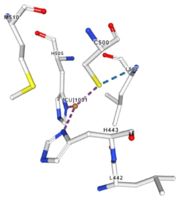
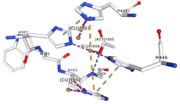
</td></tr><tr><td>[[Image:Cu1001.jpg|thumb|left|'''Figure 2:''' Copper 1001<ref>RCSB PDB</ref>]]</td><td>Cu1001: The copper ion is bound thanks to 3 metal protein interactions with H443, H505 and C500, structure stabilised by L502, M510 by hydrogen bonds</td></tr> | </td></tr><tr><td>[[Image:Cu1001.jpg|thumb|left|'''Figure 2:''' Copper 1001<ref>RCSB PDB</ref>]]</td><td>Cu1001: The copper ion is bound thanks to 3 metal protein interactions with H443, H505 and C500, structure stabilised by L502, M510 by hydrogen bonds</td></tr> | ||
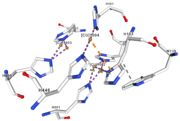
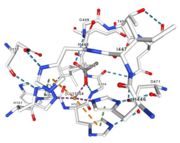
<tr><td>[[Image:Cu1002.jpg|thumb|left|'''Figure 3:''' Copper 1002<ref>RCSB PDB</ref>]]</td><td>Cu1002: 2 atoms of Cu, bonds twice by metal interaction (2x3) with 3 histidine : H501, H103, H141 stabilised by hydrophobic contact with W139</td></tr> | <tr><td>[[Image:Cu1002.jpg|thumb|left|'''Figure 3:''' Copper 1002<ref>RCSB PDB</ref>]]</td><td>Cu1002: 2 atoms of Cu, bonds twice by metal interaction (2x3) with 3 histidine : H501, H103, H141 stabilised by hydrophobic contact with W139</td></tr> | ||
| Line 78: | Line 78: | ||
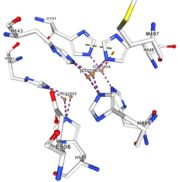
<tr><td colspan='4' class="sblockLbl">Stabilisation of Cu1002 <br> | <tr><td colspan='4' class="sblockLbl">Stabilisation of Cu1002 <br> | ||
</td></tr> | </td></tr> | ||
| - | </table> | + | </table><ref>PMID:17804014</ref> |
| + | Biochemical function: metal ion binding | ||
| + | Biological process: oxidation-reduction process | ||
| + | Cellular component: outer membrane-bounded periplasmic space | ||
| + | Sequence domains: Cupredoxin - Multicopper oxidase, type 3 - Multicopper oxidase, type 2 - Multicopper oxidase, copper-binding site - Multicopper oxidase, type 1 | ||
| + | Chain: A | ||
| + | Length: 489 amino acids | ||
| + | Theoretical weight: 53.54 KDa | ||
| + | Source organism: Escherichia coli K-12 | ||
| + | Expression system: Escherichia coli | ||
| + | UniProt: | ||
| + | Canonical: P36649 (Residues: 29-516; Coverage: 100%) | ||
| + | Gene names: JW0119, b0123, cueO, yacK | ||
| + | (Image avec flèche, image avec interactions entre résidus, image ligands environnement) | ||
| + | Validations informations (Numbers of outliers) | ||
Revision as of 06:40, 11 January 2019
Multicopper Oxidase CueO (4e9s)
| |||||||||||
References
- ↑ EMBL-EBI, Family: Cu-oxidase (PF00394), Summary: Multicopper oxidase, http://pfam.xfam.org/family/Cu-oxidase
- ↑ <ref>PMID:9556453</ref>
- ↑ <ref>PMID:9556453</ref>
- ↑ <ref>PMID:9556453</ref>
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ RCSB PDB
- ↑ Kataoka K, Komori H, Ueki Y, Konno Y, Kamitaka Y, Kurose S, Tsujimura S, Higuchi Y, Kano K, Seo D, Sakurai T. Structure and function of the engineered multicopper oxidase CueO from Escherichia coli--deletion of the methionine-rich helical region covering the substrate-binding site. J Mol Biol. 2007 Oct 12;373(1):141-52. Epub 2007 Aug 2. PMID:17804014 doi:10.1016/j.jmb.2007.07.041